A novel method for finding tRNA genes
- PMID: 12702810
- PMCID: PMC1370417
- DOI: 10.1261/rna.2193703
A novel method for finding tRNA genes
Abstract
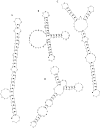
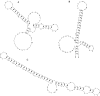
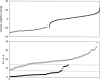
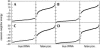
We describe a novel procedure for generating and optimizing pattern descriptors that can be used to find structural motifs in DNA or RNA sequences. This combines a pattern-description language (based primarily on secondary structure alignment and conservation of some key nucleotides) with a scoring function that relies heavily on estimated folding free energies for the secondary structure of interest. For the cloverleaf secondary structure characteristic of tRNA, we show that a fairly simple pattern descriptor can find almost all known tRNA genes in both bacterial and eukaryotic genomes, and that false positives (sequences that match the pattern but that are probably not tRNAs) can be recognized by their high estimated folding free energies. A general procedure for optimizing descriptors (and hence for finding new structural motifs) is also described. For six bacterial, four eukaryotic, and four archaea genome sequences, our results compare favorably with those of the more complex and specialized tRNAscan-SE algorithm. Prospects for using this general approach to find other RNA structural motifs are discussed.
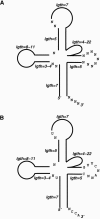
Figures
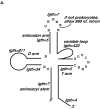

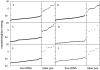
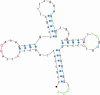
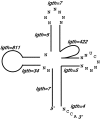

Similar articles
-
ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences.Nucleic Acids Res. 2004 Jan 2;32(1):11-6. doi: 10.1093/nar/gkh152. Print 2004. Nucleic Acids Res. 2004. PMID: 14704338 Free PMC article.
-
Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales.Mol Biol Evol. 2008 Dec;25(12):2709-16. doi: 10.1093/molbev/msn216. Epub 2008 Oct 1. Mol Biol Evol. 2008. PMID: 18832079
-
SPLITS: a new program for predicting split and intron-containing tRNA genes at the genome level.In Silico Biol. 2006;6(5):411-8. In Silico Biol. 2006. PMID: 17274770
-
Dimer formation by tRNAs.Biochemistry (Mosc). 1999 Mar;64(3):298-306. Biochemistry (Mosc). 1999. PMID: 10205299 Review.
-
[The tRNA genes of pro- and eukaryotes. Organization, structure, transcription].Mol Biol (Mosk). 1985 Jan-Feb;19(1):36-54. Mol Biol (Mosk). 1985. PMID: 2580226 Review. Russian.
Cited by
-
Folding and finding RNA secondary structure.Cold Spring Harb Perspect Biol. 2010 Dec;2(12):a003665. doi: 10.1101/cshperspect.a003665. Epub 2010 Aug 4. Cold Spring Harb Perspect Biol. 2010. PMID: 20685845 Free PMC article. Review.
-
RNAPattMatch: a web server for RNA sequence/structure motif detection based on pattern matching with flexible gaps.Nucleic Acids Res. 2015 Jul 1;43(W1):W507-12. doi: 10.1093/nar/gkv435. Epub 2015 May 4. Nucleic Acids Res. 2015. PMID: 25940619 Free PMC article.
-
ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences.Nucleic Acids Res. 2004 Jan 2;32(1):11-6. doi: 10.1093/nar/gkh152. Print 2004. Nucleic Acids Res. 2004. PMID: 14704338 Free PMC article.
-
Fluorescence competition and optical melting measurements of RNA three-way multibranch loops provide a revised model for thermodynamic parameters.Biochemistry. 2011 Feb 8;50(5):640-53. doi: 10.1021/bi101470n. Epub 2011 Jan 14. Biochemistry. 2011. PMID: 21133351 Free PMC article.
-
Searching for IRES.RNA. 2006 Oct;12(10):1755-85. doi: 10.1261/rna.157806. Epub 2006 Sep 6. RNA. 2006. PMID: 16957278 Free PMC article. Review.
References
-
- Deutscher, M.P. 1982. tRNA nucleotidyltransferase. The Enzymes 15: 183–215.
-
- Diamond, J.M., Turner, D.H., and Mathews, D.H. 2001. Thermodynamics of three-way multibranch loops in RNA. Biochemistry 40: 6971–6981. - PubMed
-
- Durbin, R., Eddy, S., Krogh, A., and Mitchison, G. 1998. Biological sequence analysis. Probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, UK.
-
- Fichant, G.A. and Burks, C. 1991. Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol. 220: 659–671. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
