Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells
- PMID: 12697816
- PMCID: PMC153196
- DOI: 10.1128/MCB.23.9.3152-3162.2003
Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells
Abstract
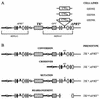
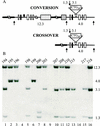
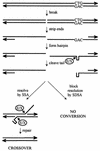
Expansion of CTG triplet repeats in the 3' untranslated region of the DMPK gene causes the autosomal dominant disorder myotonic dystrophy. Instability of CTG repeats is thought to arise from their capacity to form hairpin DNA structures. How these structures interact with various aspects of DNA metabolism has been studied intensely for Escherichia coli and Saccharomyces cerevisiae but is relatively uncharacterized in mammalian cells. To examine the stability of (CTG)(17), (CTG)(98), and (CTG)(183) repeats during homologous recombination, we placed them in the second intron of one copy of a tandemly duplicated pair of APRT genes. Cells selected for homologous recombination between the two copies of the APRT gene displayed distinctive patterns of change. Among recombinants from cells with (CTG)(98) and (CTG)(183), 5% had lost large numbers of repeats and 10% had suffered rearrangements, a frequency more than 50-fold above normal levels. Analysis of individual rearrangements confirmed the involvement of the CTG repeats. Similar changes were not observed in proliferating (CTG)(98) and (CTG)(183) cells that were not recombinant at APRT. Instead, they displayed high frequencies of small changes in repeat number. The (CTG)(17) repeats were stable in all assays. These studies indicate that homologous recombination strongly destabilizes long tracts of CTG repeats.
Figures


Similar articles
-
A novel selectable system for detecting expansion of CAG.CTG repeats in mammalian cells.Mutat Res. 2005 May 2;572(1-2):123-31. doi: 10.1016/j.mrfmmm.2005.01.013. Mutat Res. 2005. PMID: 15790495
-
Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells.Mol Cell Biol. 2003 Jul;23(13):4485-93. doi: 10.1128/MCB.23.13.4485-4493.2003. Mol Cell Biol. 2003. PMID: 12808091 Free PMC article.
-
Long CTG.CAG repeat sequences markedly stimulate intramolecular recombination.J Biol Chem. 2002 Sep 13;277(37):34087-100. doi: 10.1074/jbc.M202128200. Epub 2002 Jun 3. J Biol Chem. 2002. PMID: 12045198
-
Myotonic dystrophy: clinical and molecular parallels between myotonic dystrophy type 1 and type 2.Curr Neurol Neurosci Rep. 2002 Sep;2(5):465-70. doi: 10.1007/s11910-002-0074-6. Curr Neurol Neurosci Rep. 2002. PMID: 12169228 Review.
-
Myotonic dystrophy: an unstable CTG repeat in a protein kinase gene.Semin Cell Biol. 1995 Feb;6(1):13-9. doi: 10.1016/1043-4682(95)90010-1. Semin Cell Biol. 1995. PMID: 7620117 Review.
Cited by
-
Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region.Nucleic Acids Res. 2006 May 19;34(9):2663-75. doi: 10.1093/nar/gkl354. Print 2006. Nucleic Acids Res. 2006. PMID: 16714445 Free PMC article.
-
Double-strand breaks in the myotonic dystrophy type 1 and the fragile X syndrome triplet repeat sequences induce different types of mutations in DNA flanking sequences in Escherichia coli.Nucleic Acids Res. 2006;34(19):5369-82. doi: 10.1093/nar/gkl612. Epub 2006 Sep 29. Nucleic Acids Res. 2006. PMID: 17012280 Free PMC article.
-
Genome-wide demethylation promotes triplet repeat instability independently of homologous recombination.DNA Repair (Amst). 2008 Feb 1;7(2):313-20. doi: 10.1016/j.dnarep.2007.11.002. DNA Repair (Amst). 2008. PMID: 18083071 Free PMC article.
-
DNA base excision repair: a mechanism of trinucleotide repeat expansion.Trends Biochem Sci. 2012 Apr;37(4):162-72. doi: 10.1016/j.tibs.2011.12.002. Epub 2012 Jan 27. Trends Biochem Sci. 2012. PMID: 22285516 Free PMC article. Review.
-
Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells.Hum Mol Genet. 2013 Dec 20;22(25):5276-87. doi: 10.1093/hmg/ddt386. Epub 2013 Aug 9. Hum Mol Genet. 2013. PMID: 23933738 Free PMC article.
References
-
- Ashizawa, T., D. G. Monckton, S. Vaishnav, B. J. Patel, A. Voskova, and C. T. Caskey. 1996. Instability of the expanded (CTG)n repeats in the myotonin protein kinase gene in cultured lymphoblastoid cell lines from patients with myotonic dystrophy. Genomics 36:47-53. - PubMed
-
- Ashley, C. T., and S. T. Warren. 1995. Trinucleotide repeat expansion and human disease. Annu. Rev. Genet. 29:703-728. - PubMed
-
- Balakumaran, B. S., C. H. Freudenreich, and V. A. Zakian. 2000. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 9:93-100. - PubMed
-
- Benson, F. E., P. Baumann, and S. C. West. 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391:401-404. - PubMed
-
- Bollag, R. J., A. S. Waldman, and R. M. Liskay. 1989. Homologous recombination in mammalian cells. Annu. Rev. Genet. 23:199-225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
