Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells
- PMID: 12692243
- PMCID: PMC153947
- DOI: 10.1128/jvi.77.9.5415-5427.2003
Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells
Abstract
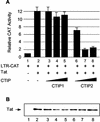
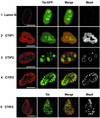
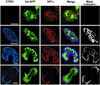
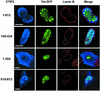
The Tat protein of human immunodeficiency virus type 1 (HIV-1) plays a key role as inducer of viral gene expression. We report that Tat function can be potently inhibited in human microglial cells by the recently described nuclear receptor cofactor chicken ovalbumin upstream promoter transcription factor-interacting protein 2 (CTIP2). Overexpression of CTIP2 leads to repression of HIV-1 replication, as a result of inhibition of Tat-mediated transactivation. In contrast, the related CTIP1 was unable to affect Tat function and viral replication. Using confocal microscopy to visualize Tat subcellular distribution in the presence of the CTIPs, we found that overexpression of CTIP2, and not of CTIP1, leads to disruption of Tat nuclear localization and recruitment of Tat within CTIP2-induced nuclear ball-like structures. In addition, our studies demonstrate that CTIP2 colocalizes and associates with the heterochromatin-associated protein HP1alpha. The CTIP2 protein harbors two Tat and HP1 interaction interfaces, the 145-434 and the 717-813 domains. CTIP2 and HP1alpha associate with Tat to form a three-protein complex in which the 145-434 CTIP2 domain interacts with the N-terminal region of Tat, while the 717-813 domain binds to HP1. The importance of this Tat binding interface and of Tat subnuclear relocation was confirmed by analysis of CTIP2 deletion mutants. Our findings suggest that inhibition of HIV-1 expression by CTIP2 correlates with recruitment of Tat within CTIP2-induced structures and relocalization within inactive regions of the chromatin via formation of the Tat-CTIP2-HP1alpha complex. These data highlight a new mechanism of Tat inactivation through subnuclear relocalization that may ultimately lead to inhibition of viral pathogenesis.
Figures




Similar articles
-
COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells.Nucleic Acids Res. 2005 Apr 22;33(7):2318-31. doi: 10.1093/nar/gki529. Print 2005. Nucleic Acids Res. 2005. Retraction in: Nucleic Acids Res. 2023 May 09:gkad358. doi: 10.1093/nar/gkad358 PMID: 15849318 Free PMC article. Retracted.
-
The nuclear receptor chicken ovalbumin upstream promoter transcription factor interacts with HIV-1 Tat and stimulates viral replication in human microglial cells.J Biol Chem. 2000 Jan 28;275(4):2654-60. doi: 10.1074/jbc.275.4.2654. J Biol Chem. 2000. PMID: 10644726
-
Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression.J Biol Chem. 2003 Oct 31;278(44):43041-50. doi: 10.1074/jbc.M307477200. Epub 2003 Aug 19. J Biol Chem. 2003. PMID: 12930829 Free PMC article.
-
Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
-
Multiple actions of the human immunodeficiency virus type-1 Tat protein on microglial cell functions.Neurochem Res. 2004 May;29(5):965-78. doi: 10.1023/b:nere.0000021241.90133.89. Neurochem Res. 2004. PMID: 15139295 Review.
Cited by
-
Microglial Cells: The Main HIV-1 Reservoir in the Brain.Front Cell Infect Microbiol. 2019 Oct 24;9:362. doi: 10.3389/fcimb.2019.00362. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31709195 Free PMC article. Review.
-
COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells.Nucleic Acids Res. 2005 Apr 22;33(7):2318-31. doi: 10.1093/nar/gki529. Print 2005. Nucleic Acids Res. 2005. Retraction in: Nucleic Acids Res. 2023 May 09:gkad358. doi: 10.1093/nar/gkad358 PMID: 15849318 Free PMC article. Retracted.
-
Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing.EMBO J. 2007 Jan 24;26(2):412-23. doi: 10.1038/sj.emboj.7601516. EMBO J. 2007. PMID: 17245431 Free PMC article.
-
Curing HIV: Pharmacologic approaches to target HIV-1 latency.Annu Rev Pharmacol Toxicol. 2011;51:397-418. doi: 10.1146/annurev-pharmtox-010510-100237. Annu Rev Pharmacol Toxicol. 2011. PMID: 21210747 Free PMC article. Review.
-
HIV Associated Neurocognitive Disorders.Infect Dis Rep. 2013 Jun 6;5(Suppl 1):e8. doi: 10.4081/idr.2013.s1.e8. eCollection 2013 Jun 6. Infect Dis Rep. 2013. PMID: 24470972 Free PMC article. Review.
References
-
- Avram, D., A. Fields, K. Pretty On Top, D. J. Nevrivy, J. E. Ishmael, and M. Leid. 2000. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 275:10315-10322. - PMC - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Benkirane, M., R. F. Chun, H. Xiao, V. V. Ogryzko, B. H. Howard, Y. Nakatani, and K. T. Jeang. 1998. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 273:24898-24905. - PubMed
-
- Boykins, R. A., R. Mahieux, U. T. Shankavaram, Y. S. Gho, S. F. Lee, I. K. Hewlett, L. M. Wahl, H. K. Kleinman, J. N. Brady, K. M. Yamada, and S. Dhawan. 1999. Cutting edge: a short polypeptide domain of HIV-1-Tat protein mediates pathogenesis. J. Immunol. 163:15-20. - PubMed
-
- Brasher, S. V., B. O. Smith, R. H. Fogh, D. Nietlispach, A. Thiru, P. R. Nielsen, R. W. Broadhurst, L. J. Ball, N. V. Murzina, and E. D. Laue. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19:1587-1597. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

