Replication studies using genotype 1a subgenomic hepatitis C virus replicons
- PMID: 12692237
- PMCID: PMC153987
- DOI: 10.1128/jvi.77.9.5352-5359.2003
Replication studies using genotype 1a subgenomic hepatitis C virus replicons
Abstract
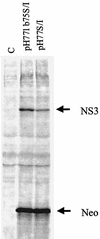
Recently, cell-based replicon systems for hepatitis C virus (HCV), in which the nonstructural proteins stably replicate subgenomic viral RNA in Huh7 cells, were developed. To date, one limitation of using these replicon systems to advance drug discovery is the inability of other genotypic derivatives, beyond those of two distinct strains of genotype 1b (HCV-N and Con1), to stably replicate in Huh7 cells. In this report, we evaluated a series of replicon genotype 1a-1b chimeras, as well as a complete genotype 1a replicon clone. A subgenomic replicon construct containing only type 1a sequences failed to generate stable colonies in Huh7 cells even after repeated attempts. Furthermore, addition of an NS5A adaptive mutation (S2204I) which enhances type 1b replicon efficiency was insufficient to confer replication to the wild-type 1a replicon. This subgenomic replicon was subsequently found to be inefficiently translated in Huh7 cells compared to a type 1b replicon, and the attenuation of translation mapped to the N-terminal region of NS3. Therefore, to ensure efficient translation and thereby support replication of the 1a genome, the coding sequence for first 75 residues from type 1a were replaced with the type 1b (strain Con 1) NS3 coding sequence. Although nonstructural proteins were expressed at lower levels with this replicon than with type 1b and although the amount of viral RNA was also severalfold lower (150 copies of positive-strand RNA per cell), the replicon stably replicated in Huh7 cells. Notwithstanding this difference, the ratio of positive- to negative-strand RNA of 26 was similar to that found with the type 1b replicon. Similar results were found for a 1b replicon expressing the type 1a RNA-dependent RNA polymerase. These 1a hybrid replicons maintained sensitivity to alpha interferon (IFN-alpha), albeit with an eightfold-higher 50% inhibitory concentration than type 1b replicons. Evidence is provided herein to confirm that this differential response to IFN-alpha may be attributed directly to the type 1a polymerase.
Figures



Similar articles
-
Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons.J Virol. 2003 Jan;77(2):1092-104. doi: 10.1128/jvi.77.2.1092-1104.2003. J Virol. 2003. PMID: 12502825 Free PMC article.
-
Novel hepatitis C virus reporter replicon cell lines enable efficient antiviral screening against genotype 1a.Antimicrob Agents Chemother. 2010 Aug;54(8):3099-106. doi: 10.1128/AAC.00289-10. Epub 2010 Jun 1. Antimicrob Agents Chemother. 2010. PMID: 20516274 Free PMC article.
-
Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells.J Virol. 2002 Mar;76(6):2997-3006. doi: 10.1128/jvi.76.6.2997-3006.2002. J Virol. 2002. PMID: 11861865 Free PMC article.
-
Replication of the hepatitis C virus in cell culture.Antiviral Res. 2003 Oct;60(2):91-102. doi: 10.1016/j.antiviral.2003.08.016. Antiviral Res. 2003. PMID: 14638404 Review.
-
[Interferon resistance and ISDR (interferon sensitivity determining region)].Nihon Rinsho. 2006 Jul;64(7):1249-53. Nihon Rinsho. 2006. PMID: 16838640 Review. Japanese.
Cited by
-
Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes.J Virol. 2004 Jul;78(14):7400-9. doi: 10.1128/JVI.78.14.7400-7409.2004. J Virol. 2004. PMID: 15220413 Free PMC article.
-
Genetic interactions between hepatitis C virus replicons.J Virol. 2004 Nov;78(21):12085-9. doi: 10.1128/JVI.78.21.12085-12089.2004. J Virol. 2004. PMID: 15479852 Free PMC article.
-
Hepatitis C virus induced a novel apoptosis-like death of pancreatic beta cells through a caspase 3-dependent pathway.PLoS One. 2012;7(6):e38522. doi: 10.1371/journal.pone.0038522. Epub 2012 Jun 4. PLoS One. 2012. PMID: 22675572 Free PMC article.
-
Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells.J Virol. 2004 Aug;78(15):7904-15. doi: 10.1128/JVI.78.15.7904-7915.2004. J Virol. 2004. PMID: 15254163 Free PMC article.
-
Secondary Structural Elements of the HCV X-region Involved in Viral Replication.J Clin Transl Hepatol. 2015 Mar;3(1):1-8. doi: 10.14218/JCTH.2015.00003. Epub 2015 Mar 15. J Clin Transl Hepatol. 2015. PMID: 26356238 Free PMC article.
References
-
- Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. - PubMed
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Brass, V., E. Bieck, R. Montserret, V. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. - PubMed
-
- Craggs, J., J. Ball, B. Thomson, W. Irving, and A. Grabowska. 2001. Development of a strand-specific RT-PCR based assay to detect the replicative form of hepatitis C virus RNA. J. Virol. Methods 94:111-120. - PubMed
-
- Dhanak, D., K. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. G. B. Shaw, C. Silverman, A. T. Gates, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H.-Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

