Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line
- PMID: 12660171
- PMCID: PMC152905
- DOI: 10.1093/emboj/cdg161
Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line
Abstract
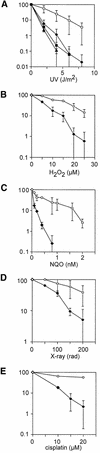
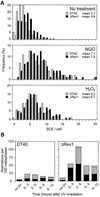
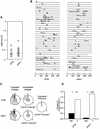
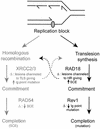
The majority of DNA damage-induced mutagenesis in the yeast Saccharomyces cerevisiae arises as a result of translesion replication. This process is critically dependent on the deoxycytidyl transferase Rev1p, which forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p. To examine the role of Rev1 in vertebrate mutagenesis and the DNA damage response, we disrupted the gene in DT40 cells. Rev1-deficient DT40 grow slowly and are sensitive to a wide range of DNA-damaging agents. Homologous recombination repair is likely to be intact as basal and damage induced sister chromatid exchange and immunoglobulin gene conversion are unaffected. How ever, the mutant cells show a markedly reduced level of non-templated immunoglobulin gene mutation, indicating a defect in translesion bypass. Furthermore, ultraviolet exposure results in marked chromosome breakage, suggesting that replication gaps created in the absence of Rev1 cannot be efficiently repaired by recombination. Thus, Rev1-dependent translesion bypass and mutagenesis is likely to be a trade-off for the ability to complete replication of a damaged template and thereby maintain genome integrity.
Figures

Similar articles
-
The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40.Mol Immunol. 2006 Apr;43(10):1587-94. doi: 10.1016/j.molimm.2005.09.017. Epub 2005 Nov 2. Mol Immunol. 2006. PMID: 16263170
-
UBE2V2 (MMS2) is not required for effective immunoglobulin gene conversion or DNA damage tolerance in DT40.DNA Repair (Amst). 2005 Apr 4;4(4):503-10. doi: 10.1016/j.dnarep.2004.12.002. Epub 2005 Jan 21. DNA Repair (Amst). 2005. PMID: 15725630
-
A non-catalytic function of Rev1 in translesion DNA synthesis and mutagenesis is mediated by its stable interaction with Rad5.DNA Repair (Amst). 2013 Jan 1;12(1):27-37. doi: 10.1016/j.dnarep.2012.10.003. Epub 2012 Nov 9. DNA Repair (Amst). 2013. PMID: 23142547
-
Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p.Philos Trans R Soc Lond B Biol Sci. 2001 Jan 29;356(1405):41-6. doi: 10.1098/rstb.2000.0001. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11205328 Free PMC article. Review.
-
[Induced mutagenesis and translesion DNA synthesis--structure and function of REV1].Seikagaku. 2008 Sep;80(9):843-6. Seikagaku. 2008. PMID: 18975621 Review. Japanese. No abstract available.
Cited by
-
Translesion DNA synthesis and mutagenesis in eukaryotes.Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a012708. doi: 10.1101/cshperspect.a012708. Cold Spring Harb Perspect Biol. 2013. PMID: 23457261 Free PMC article. Review.
-
Ancient phylogenetic beginnings of immunoglobulin hypermutation.J Mol Evol. 2006 Nov;63(5):691-706. doi: 10.1007/s00239-006-0051-9. Epub 2006 Oct 6. J Mol Evol. 2006. PMID: 17031458
-
DNA-dependent protein kinase inhibits AID-induced antibody gene conversion.PLoS Biol. 2007 Apr;5(4):e80. doi: 10.1371/journal.pbio.0050080. PLoS Biol. 2007. PMID: 17355182 Free PMC article.
-
REV1 accumulates in DNA damage-induced nuclear foci in human cells and is implicated in mutagenesis by benzo[a]pyrenediolepoxide.Nucleic Acids Res. 2004 Nov 2;32(19):5820-6. doi: 10.1093/nar/gkh903. Print 2004. Nucleic Acids Res. 2004. PMID: 15523096 Free PMC article.
-
Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair.Mol Cell. 2014 May 8;54(3):472-84. doi: 10.1016/j.molcel.2014.03.014. Epub 2014 Apr 10. Mol Cell. 2014. PMID: 24726326 Free PMC article.
References
-
- Arakawa H., Hauschild,J. and Buerstedde,J.M. (2002) Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science, 295, 1301–1306. - PubMed
-
- Bemark M., Khamlichi,A.A., Davies,S.L. and Neuberger,M.S. (2000) Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol., 10, 1213–1216. - PubMed
-
- Bezzubova O., Silbergleit,A., Yamaguchi-Iwai,Y., Takeda,S. and Buerstedde,J.M. (1997) Reduced X-ray resistance and homologous recombination frequencies in a RAD54–/– mutant of the chicken DT40 cell line. Cell, 89, 185–193. - PubMed
-
- Broomfield S., Hryciw,T. and Xiao,W. (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res., 486, 167–184. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

