Protein motion from non-specific to specific DNA by three-dimensional routes aided by supercoiling
- PMID: 12628933
- PMCID: PMC151056
- DOI: 10.1093/emboj/cdg125
Protein motion from non-specific to specific DNA by three-dimensional routes aided by supercoiling
Abstract
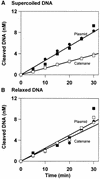
DNA-binding proteins are generally thought to locate their target sites by first associating with the DNA at random and then translocating to the specific site by one-dimensional (1D) diffusion along the DNA. We report here that non-specific DNA conveys proteins to their target sites just as well when held near the target by catenation as when co-linear with the target. Hence, contrary to the prevalent view, proteins move from random to specific sites primarily by three-dimensional (3D) rather than 1D pathways, by multiple dissociation/re-association events within a single DNA molecule. We also uncover a role for DNA supercoiling in target-site location. Proteins find their sites more readily in supercoiled than in relaxed DNA, again indicating 3D rather than 1D routes.
Figures
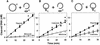

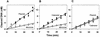
Similar articles
-
How to get from A to B: strategies for analysing protein motion on DNA.Eur Biophys J. 2002 Jul;31(4):257-67. doi: 10.1007/s00249-002-0224-4. Epub 2002 May 30. Eur Biophys J. 2002. PMID: 12122472 Review.
-
Communications between distant sites on supercoiled DNA from non-exponential kinetics for DNA synapsis by resolvase.J Mol Biol. 1997 Jul 18;270(3):396-412. doi: 10.1006/jmbi.1997.1109. J Mol Biol. 1997. PMID: 9237906
-
Interactions of protein complexes on supercoiled DNA: the mechanism of selective synapsis by Tn3 resolvase.J Mol Biol. 2002 May 31;319(2):371-83. doi: 10.1016/S0022-2836(02)00309-1. J Mol Biol. 2002. PMID: 12051914
-
DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes.EMBO J. 1999 May 4;18(9):2638-47. doi: 10.1093/emboj/18.9.2638. EMBO J. 1999. PMID: 10228175 Free PMC article.
-
An end to 40 years of mistakes in DNA-protein association kinetics?Biochem Soc Trans. 2009 Apr;37(Pt 2):343-8. doi: 10.1042/BST0370343. Biochem Soc Trans. 2009. PMID: 19290859 Review.
Cited by
-
Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria.Proc Natl Acad Sci U S A. 2012 May 15;109(20):E1287-93. doi: 10.1073/pnas.1119226109. Epub 2012 Apr 16. Proc Natl Acad Sci U S A. 2012. PMID: 22509013 Free PMC article.
-
Zero mode waveguides for single-molecule spectroscopy on lipid membranes.Biophys J. 2006 May 1;90(9):3288-99. doi: 10.1529/biophysj.105.072819. Epub 2006 Feb 3. Biophys J. 2006. PMID: 16461393 Free PMC article.
-
DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway.Nucleic Acids Res. 2005 May 10;33(8):2676-84. doi: 10.1093/nar/gki565. Print 2005. Nucleic Acids Res. 2005. PMID: 15886396 Free PMC article.
-
Central dogma at the single-molecule level in living cells.Nature. 2011 Jul 20;475(7356):308-15. doi: 10.1038/nature10315. Nature. 2011. PMID: 21776076 Free PMC article. Review.
-
Rules of engagement for base excision repair in chromatin.J Cell Physiol. 2013 Feb;228(2):258-66. doi: 10.1002/jcp.24134. J Cell Physiol. 2013. PMID: 22718094 Free PMC article. Review.
References
-
- Adzuma K. and Mizuuchi,K. (1989) Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell, 57, 41–47. - PubMed
-
- Amouyal M. and Buc,H. (1987) Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and UV5 sites of Escherichia coli. J. Mol. Biol., 195, 795–808. - PubMed
-
- Baldwin G.S., Sessions,R.B., Erskine,S.G. and Halford,S.E. (1999) DNA cleavage by the EcoRV restriction endonuclease: roles of divalent metal ions in specificity and catalysis. J. Mol. Biol., 288, 87–103. - PubMed
-
- Bates A.D. and Maxwell,A. (1993) DNA Topology. IRL Press, Oxford, UK.
-
- Berg H.C. (1993) Random Walks in Biology. Princeton University Press, Princeton, NJ.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

