Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors
- PMID: 12612064
- PMCID: PMC149485
- DOI: 10.1128/MCB.23.6.1885-1895.2003
Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors
Abstract
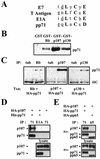
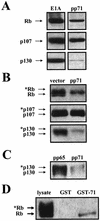
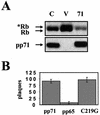
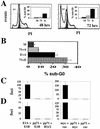
The oncoproteins of the DNA tumor viruses, adenovirus E1A, simian virus 40 T antigen, and papillomavirus E7, each interact with the retinoblastoma family of tumor suppressors, leading to cell cycle stimulation, apoptosis induction, and cellular transformation. These proteins utilize a conserved LXCXE motif, which is also found in cellular proteins, to target the retinoblastoma family. Here, we describe a herpesvirus protein that shares a subset of the properties of the DNA tumor virus oncoproteins but maintains important differences as well. The human cytomegalovirus pp71 protein employs an LXCXD motif to attack the retinoblastoma family members and induce DNA synthesis in quiescent cells. pp71 binds to and induces the degradation of the hypophosphorylated forms of the retinoblastoma protein and its family members p107 and p130 in a proteasome-dependent manner. However, pp71 does not induce apoptosis and fails to transform cells. Thus, the similarities and differences in comparison to E1A, T antigen, and E7 make pp71 an interesting new tool with which to further dissect the role of the retinoblastoma/E2F pathway in cellular growth control and carcinogenesis.
Figures




Similar articles
-
Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein.Proc Natl Acad Sci U S A. 2003 Mar 18;100(6):3263-8. doi: 10.1073/pnas.0538058100. Epub 2003 Mar 7. Proc Natl Acad Sci U S A. 2003. PMID: 12626766 Free PMC article.
-
E2F-Rb complexes assemble and inhibit cdc25A transcription in cervical carcinoma cells following repression of human papillomavirus oncogene expression.Mol Cell Biol. 2000 Oct;20(19):7059-67. doi: 10.1128/MCB.20.19.7059-7067.2000. Mol Cell Biol. 2000. PMID: 10982822 Free PMC article.
-
Human cytomegalovirus pp71: a new viral tool to probe the mechanisms of cell cycle progression and oncogenesis controlled by the retinoblastoma family of tumor suppressors.J Cell Biochem. 2004 Sep 1;93(1):37-45. doi: 10.1002/jcb.20177. J Cell Biochem. 2004. PMID: 15352160 Review.
-
E1A 12S and 13S of the transformation-defective adenovirus type 12 strain CS-1 inactivate proteins of the RB family, permitting transactivation of the E2F-dependent promoter.J Virol. 1997 Dec;71(12):9538-48. doi: 10.1128/JVI.71.12.9538-9548.1997. J Virol. 1997. PMID: 9371617 Free PMC article.
-
Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation.Crit Rev Biochem Mol Biol. 1996 Jun;31(3):237-71. doi: 10.3109/10409239609106585. Crit Rev Biochem Mol Biol. 1996. PMID: 8817077 Review.
Cited by
-
Protein S-Nitrosylation of Human Cytomegalovirus pp71 Inhibits Its Ability To Limit STING Antiviral Responses.J Virol. 2020 Aug 17;94(17):e00033-20. doi: 10.1128/JVI.00033-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581105 Free PMC article.
-
Proteasome-dependent disruption of the E3 ubiquitin ligase anaphase-promoting complex by HCMV protein pUL21a.PLoS Pathog. 2012;8(7):e1002789. doi: 10.1371/journal.ppat.1002789. Epub 2012 Jul 5. PLoS Pathog. 2012. PMID: 22792066 Free PMC article.
-
Virus-host protein interactions as footprints of human cytomegalovirus replication.Curr Opin Virol. 2022 Feb;52:135-147. doi: 10.1016/j.coviro.2021.11.016. Epub 2021 Dec 16. Curr Opin Virol. 2022. PMID: 34923282 Free PMC article. Review.
-
The human cytomegalovirus UL26 protein antagonizes NF-κB activation.J Virol. 2014 Dec;88(24):14289-300. doi: 10.1128/JVI.02552-14. Epub 2014 Oct 1. J Virol. 2014. PMID: 25275128 Free PMC article.
-
Cell cycle regulation during viral infection.Methods Mol Biol. 2014;1170:165-227. doi: 10.1007/978-1-4939-0888-2_10. Methods Mol Biol. 2014. PMID: 24906315 Free PMC article. Review.
References
-
- Berezutskaya, E., B. Yu, A. Morozov, P. Raychaudhuri, and S. Bagchi. 1997. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 8:1277-1286. - PubMed
-
- Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papillomavirus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. - PubMed
-
- Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in G1. Virology 224:150-160. - PubMed
-
- Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
