An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis
- PMID: 12584345
- PMCID: PMC149741
- DOI: 10.1128/jvi.77.5.3217-3228.2003
An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis
Abstract
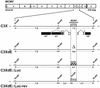
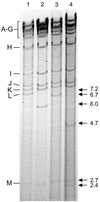
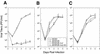
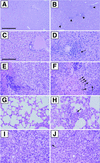
The transcription of cytomegalovirus (CMV) immediate-early (IE) genes is regulated by a large and complex enhancer containing an array of binding sites for a variety of cellular transcription factors. Previously, using bacterial artificial chromosome recombinants of the virus genome, it was reported that the enhancer region of murine CMV (MCMV) is dispensable but performs a key function for viral multiplication (A. Angulo, M. Messerle, U. H. Koszinowski, and P. Ghazal, J. Virol. 72:8502-8509, 1998). In the present study, we defined, through the reconstitution of infectious enhancerless MCMVs, the growth requirement for the enhancer in tissue culture and explored its significance for steering a productive infection in vivo. A comparison of cis and trans complementation systems for infection of enhancerless virus in permissive fibroblasts revealed a multiplicity-dependent growth phenotype that is severely compromised in the rate of infectious-virus multiplication. The in vivo impact of viruses that have an amputated enhancer was investigated in an extremely sensitive model of MCMV infection, the SCID mouse. Histological examination of spleens, livers, lungs, and salivary glands from animals infected with enhancer-deficient MCMV demonstrated an absence of tissue damage associated with CMV infection. The lack of pathogenic lesions correlated with a defect in replication competence. Enhancerless viruses were not detectable in major target organs harvested from SCID mice. The pathogenesis and growth defect reverted upon restoration of the enhancer. Markedly, while SCID mice infected with 5 PFU of parental MCMV died within 50 days postinfection, all mice infected with enhancerless virus survived for the duration of the experiment (1 year) after infection with 5 x 10(5) PFU. Together, these results clarify the importance of the enhancer for MCMV growth in cell culture and underscore the in vivo significance of this region for MCMV virulence and pathogenesis.
Figures



Similar articles
-
Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture.J Virol. 2005 Jun;79(11):7182-94. doi: 10.1128/JVI.79.11.7182-7194.2005. J Virol. 2005. PMID: 15890957 Free PMC article.
-
Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer.J Virol. 1998 Nov;72(11):8502-9. doi: 10.1128/JVI.72.11.8502-8509.1998. J Virol. 1998. PMID: 9765387 Free PMC article.
-
Murine cytomegalovirus with a transposon insertional mutation at open reading frame m155 is deficient in growth and virulence in mice.J Virol. 2004 Jul;78(13):6891-9. doi: 10.1128/JVI.78.13.6891-6899.2004. J Virol. 2004. PMID: 15194765 Free PMC article.
-
Genetic analyses of gene function and pathogenesis of murine cytomegalovirus by transposon-mediated mutagenesis.J Clin Virol. 2002 Aug;25 Suppl 2:S111-22. doi: 10.1016/s1386-6532(02)00096-3. J Clin Virol. 2002. PMID: 12361762 Review.
-
Conditional gene expression systems to study herpesvirus biology in vivo.Med Microbiol Immunol. 2008 Jun;197(2):269-76. doi: 10.1007/s00430-008-0086-1. Epub 2008 Mar 7. Med Microbiol Immunol. 2008. PMID: 18324415 Review.
Cited by
-
Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus.J Virol. 2007 Dec;81(24):13761-70. doi: 10.1128/JVI.01290-07. Epub 2007 Oct 10. J Virol. 2007. PMID: 17928340 Free PMC article.
-
Activation of hepatic natural killer cells and control of liver-adapted lymphoma in the murine model of cytomegalovirus infection.Med Microbiol Immunol. 2008 Jun;197(2):167-78. doi: 10.1007/s00430-008-0084-3. Epub 2008 Feb 29. Med Microbiol Immunol. 2008. PMID: 18309517 Review.
-
Transactivation of cellular genes involved in nucleotide metabolism by the regulatory IE1 protein of murine cytomegalovirus is not critical for viral replicative fitness in quiescent cells and host tissues.J Virol. 2008 Oct;82(20):9900-16. doi: 10.1128/JVI.00928-08. Epub 2008 Aug 6. J Virol. 2008. PMID: 18684825 Free PMC article.
-
Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis.PLoS Biol. 2011 Mar;9(3):e1000598. doi: 10.1371/journal.pbio.1000598. Epub 2011 Mar 8. PLoS Biol. 2011. PMID: 21408089 Free PMC article.
-
Murine cytomegalovirus major immediate-early enhancer region operating as a genetic switch in bidirectional gene pair transcription.J Virol. 2007 Jul;81(14):7805-10. doi: 10.1128/JVI.02388-06. Epub 2007 May 9. J Virol. 2007. PMID: 17494084 Free PMC article.
References
-
- Baskar, J. F., P. P. Smith, G. Nilaver, R. A. Jupp, S. Hoffmann, N. J. Peffer, D. J. Tenney, A. M. Colberg-Poley, P. Ghazal, and J. A. Nelson. 1996. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J. Virol. 70:3207-3214. - PMC - PubMed
-
- Boshart, M., F. Weber, G. Jahn, K. Dorsch-Hasler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41:521-530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

