Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization
- PMID: 12573215
- PMCID: PMC4765734
- DOI: 10.1016/s0960-9822(03)00044-7
Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization
Abstract
Background: mRNA localization in somatic cells is an important mechanism for gene expression regulation. In fibroblasts, the protein ZBP1 associates with the sequence that localizes beta-actin mRNA to the leading edge of fibroblasts, augmenting motility. beta-actin mRNA localizes in a cytoskeleton-dependent manner, depending on intact actin and myosin ATP-hydrolysis, and is largely bound to the actin cytoskeleton. The ZBP1 protein contains four KH RNA binding domains and a classic RBD RNA binding domain. It also contains a putative nuclear import and export sequence, suggesting a nuclear phase in this protein's function.
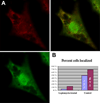
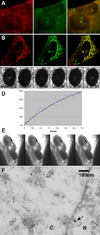
Results: Using high-speed imaging, we show here the targeting of this RNA binding protein to beta-actin pre-mRNA transcripts in the nuclei of living cells and measure the residence time of the RNA-protein complex before it leaves the transcription site. Then, the RNA-protein particle is exported to the cytoplasm, where it localizes at velocities of 0.6 microm/s by using actin filaments and/or microtubules. This RNA-ZBP1 complex is required for cytoplasmic localization in fibroblasts; mislocalizing the protein also mislocalizes the RNA, and expressing the protein in a ZBP1-deficient cell line induces beta-actin mRNA localization.
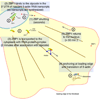
Conclusions: This work demonstrates that the RNA-protein association, essential for cytoplasmic localization, begins as soon as the RNA is transcribed. The ZBP1 then forms a ribonucleoprotein particle and moves in a myosin-dependent fashion by using the cytoskeleton for directional transport.
Figures
Similar articles
-
Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment.J Cell Biol. 2003 Jan 6;160(1):77-87. doi: 10.1083/jcb.200206003. Epub 2002 Dec 30. J Cell Biol. 2003. PMID: 12507992 Free PMC article.
-
Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1.Nature. 2005 Nov 24;438(7067):512-5. doi: 10.1038/nature04115. Nature. 2005. PMID: 16306994
-
ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription.Mol Cell Biol. 2007 Dec;27(23):8340-51. doi: 10.1128/MCB.00972-07. Epub 2007 Sep 24. Mol Cell Biol. 2007. PMID: 17893325 Free PMC article.
-
Cytoskeleton-dependent transport and localization of mRNA.Int Rev Cytol. 2001;211:1-31. doi: 10.1016/s0074-7696(01)11016-8. Int Rev Cytol. 2001. PMID: 11597002 Review.
-
How and why does beta-actin mRNA target?Biol Cell. 2005 Jan;97(1):97-110. doi: 10.1042/BC20040063. Biol Cell. 2005. PMID: 15601261 Review.
Cited by
-
Centrocortin potentiates co-translational localization of its mRNA to the centrosome via dynein.bioRxiv [Preprint]. 2024 Aug 9:2024.08.09.607365. doi: 10.1101/2024.08.09.607365. bioRxiv. 2024. PMID: 39149256 Free PMC article. Preprint.
-
VICKZ proteins mediate cell migration via their RNA binding activity.RNA. 2007 Sep;13(9):1558-69. doi: 10.1261/rna.559507. Epub 2007 Jul 24. RNA. 2007. PMID: 17652133 Free PMC article.
-
Co-transcriptional recruitment of Puf6 by She2 couples translational repression to mRNA localization.Nucleic Acids Res. 2014 Jul;42(13):8692-704. doi: 10.1093/nar/gku597. Epub 2014 Jul 10. Nucleic Acids Res. 2014. PMID: 25013181 Free PMC article.
-
Target-derived neurotrophins coordinate transcription and transport of bclw to prevent axonal degeneration.J Neurosci. 2013 Mar 20;33(12):5195-207. doi: 10.1523/JNEUROSCI.3862-12.2013. J Neurosci. 2013. PMID: 23516285 Free PMC article.
-
The zipcode-binding protein ZBP1 influences the subcellular location of the Ro 60-kDa autoantigen and the noncoding Y3 RNA.RNA. 2012 Jan;18(1):100-10. doi: 10.1261/rna.029207.111. Epub 2011 Nov 23. RNA. 2012. PMID: 22114317 Free PMC article.
References
-
- St. Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. - PubMed
-
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast con-trolled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. - PubMed
-
- Takizawa PA, Sil A, Swedlow JR, Herzkowitz I, Vale RD. Actin dependent localization of an RNA encoding a cell fate determinant in yeast. Nature. 1997;389:90–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

