Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins
- PMID: 12556496
- PMCID: PMC141151
- DOI: 10.1128/MCB.23.4.1368-1378.2003
Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins
Abstract
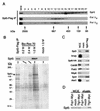
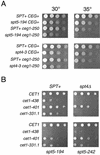
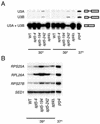
During transcription elongation, eukaryotic RNA polymerase II (Pol II) must contend with the barrier presented by nucleosomes. The conserved Spt4-Spt5 complex has been proposed to regulate elongation through nucleosomes by Pol II. To help define the mechanism of Spt5 function, we have characterized proteins that coimmunopurify with Spt5. Among these are the general elongation factors TFIIF and TFIIS as well as Spt6 and FACT, factors thought to regulate elongation through nucleosomes. Spt5 also coimmunopurified with the mRNA capping enzyme and cap methyltransferase, and spt4 and spt5 mutations displayed genetic interactions with mutations in capping enzyme genes. Additionally, we found that spt4 and spt5 mutations lead to accumulation of unspliced pre-mRNA. Spt5 also copurified with several previously unstudied proteins; we demonstrate that one of these is encoded by a new member of the SPT gene family. Finally, by immunoprecipitating these factors we found evidence that Spt5 participates in at least three Pol II complexes. These observations provide new evidence of roles for Spt4-Spt5 in pre-mRNA processing and transcription elongation.
Figures



Similar articles
-
The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation.Biochim Biophys Acta. 2013 Jan;1829(1):105-15. doi: 10.1016/j.bbagrm.2012.08.007. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982195 Free PMC article. Review.
-
Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae.Genes Dev. 1998 Feb 1;12(3):357-69. doi: 10.1101/gad.12.3.357. Genes Dev. 1998. PMID: 9450930 Free PMC article.
-
RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach.Mol Cell Biol. 2002 Oct;22(20):6979-92. doi: 10.1128/MCB.22.20.6979-6992.2002. Mol Cell Biol. 2002. PMID: 12242279 Free PMC article.
-
Biochemical Analysis of Yeast Suppressor of Ty 4/5 (Spt4/5) Reveals the Importance of Nucleic Acid Interactions in the Prevention of RNA Polymerase II Arrest.J Biol Chem. 2016 May 6;291(19):9853-70. doi: 10.1074/jbc.M116.716001. Epub 2016 Mar 4. J Biol Chem. 2016. PMID: 26945063 Free PMC article.
-
The pleiotropic roles of SPT5 in transcription.Transcription. 2022 Feb-Jun;13(1-3):53-69. doi: 10.1080/21541264.2022.2103366. Epub 2022 Jul 25. Transcription. 2022. PMID: 35876486 Free PMC article. Review.
Cited by
-
NusG-Spt5 proteins-Universal tools for transcription modification and communication.Chem Rev. 2013 Nov 13;113(11):8604-19. doi: 10.1021/cr400064k. Epub 2013 May 2. Chem Rev. 2013. PMID: 23638618 Free PMC article. Review. No abstract available.
-
The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation.Biochim Biophys Acta. 2013 Jan;1829(1):105-15. doi: 10.1016/j.bbagrm.2012.08.007. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982195 Free PMC article. Review.
-
Kcs1 and Vip1: The Key Enzymes behind Inositol Pyrophosphate Signaling in Saccharomyces cerevisiae.Biomolecules. 2024 Jan 26;14(2):152. doi: 10.3390/biom14020152. Biomolecules. 2024. PMID: 38397389 Free PMC article. Review.
-
Chromatin and transcription in yeast.Genetics. 2012 Feb;190(2):351-87. doi: 10.1534/genetics.111.132266. Genetics. 2012. PMID: 22345607 Free PMC article. Review.
-
SPT5 affects the rate of mRNA degradation and physically interacts with CCR4 but does not control mRNA deadenylation.Am J Mol Biol. 2012 Jan;2(1):11-20. doi: 10.4236/ajmb.2012.21002. Am J Mol Biol. 2012. PMID: 36910576 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Archambault, J., R. S. Chambers, M. S. Kobor, Y. Ho, M. Cartier, D. Bolotin, B. Andrews, C. M. Kane, and J. Greenblatt. 1997. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:14300-14305. - PMC - PubMed
-
- Ares, M., Jr., and A. H. Igel. 1990. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 4:2132-2145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
