Analysis of recombinant yeast decapping enzyme
- PMID: 12554866
- PMCID: PMC1370389
- DOI: 10.1261/rna.2151403
Analysis of recombinant yeast decapping enzyme
Abstract
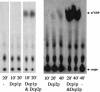
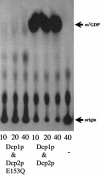
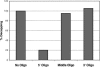
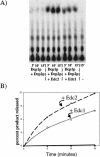
A critical step in the turnover of yeast mRNAs is decapping. Two yeast proteins, Dcp1p and Dcp2p, are absolutely required for decapping, although their precise roles in the decapping reaction have not been established. To determine the function of both Dcp1p and Dcp2p in decapping, we purified recombinant versions of these proteins from Escherichia coli and examined their properties. These experiments demonstrate that copurification of Dcp1p and Dcp2p yields active decapping enzyme under a variety of conditions. Moreover, Dcp2p alone can have decapping activity under some biochemical conditions. This suggests that Dcp2p can be a catalytic subunit of the decapping complex, and Dcp1p may function to enhance Dcp2p activity, or as an additional active subunit. In addition, recombinant Dcp1p/Dcp2p prefers long mRNA substrates and is sensitive to inhibition by sequestration of the 5' end but not the 3' end of the substrate. This suggests that Dcp1p/Dcp2p contains an additional RNA-binding site spatially distinct from the active site. Finally, using two RNA-binding proteins that enhance decapping in vivo (Edc1p and Edc2p), we can reconstitute the activation of decapping with recombinant proteins. This indicates that the Edc1 and Edc2 proteins act directly on the decapping enzyme.
Figures

Similar articles
-
Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae.Genetics. 2001 Jan;157(1):27-37. doi: 10.1093/genetics/157.1.27. Genetics. 2001. PMID: 11139489 Free PMC article.
-
The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme.RNA. 2003 Feb;9(2):239-51. doi: 10.1261/rna.2171203. RNA. 2003. PMID: 12554867 Free PMC article.
-
Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme.EMBO J. 1998 Mar 2;17(5):1487-96. doi: 10.1093/emboj/17.5.1487. EMBO J. 1998. PMID: 9482745 Free PMC article.
-
Regulation of mRNA decay: decapping goes solo.Mol Cell. 2004 Jul 2;15(1):1-2. doi: 10.1016/j.molcel.2004.06.031. Mol Cell. 2004. PMID: 15225542 Review.
-
mRNA decapping activities and their biological roles.Biochimie. 1996;78(11-12):1049-55. doi: 10.1016/s0300-9084(97)86729-6. Biochimie. 1996. PMID: 9150884 Review.
Cited by
-
Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression.Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2139-44. doi: 10.1073/pnas.0611685104. Epub 2007 Feb 5. Proc Natl Acad Sci U S A. 2007. PMID: 17283339 Free PMC article.
-
Crystal structure of Dcp1p and its functional implications in mRNA decapping.Nat Struct Mol Biol. 2004 Mar;11(3):249-56. doi: 10.1038/nsmb730. Epub 2004 Feb 1. Nat Struct Mol Biol. 2004. PMID: 14758354 Free PMC article.
-
Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms.Mol Cell. 2010 Sep 10;39(5):773-83. doi: 10.1016/j.molcel.2010.08.025. Mol Cell. 2010. PMID: 20832728 Free PMC article.
-
A quantitative assay for measuring mRNA decapping by splinted ligation reverse transcription polymerase chain reaction: qSL-RT-PCR.RNA. 2011 Mar;17(3):535-43. doi: 10.1261/rna.2436411. Epub 2011 Jan 10. RNA. 2011. PMID: 21220548 Free PMC article.
-
The control of mRNA decapping and P-body formation.Mol Cell. 2008 Dec 5;32(5):605-15. doi: 10.1016/j.molcel.2008.11.001. Mol Cell. 2008. PMID: 19061636 Free PMC article. Review.
References
-
- Beelman, C.A. and Parker, R. 1995. Degradation of mRNA in eukaryotes. Cell 8: 179–183. - PubMed
-
- Beelman, C.A., Stevens A., Caponigro G., LaGrandeur, T.E., Hatfield L., Fortner, D., and Parker, R. 1996. An essential component of the decapping enzyme required for normal rates of mRNA decay in yeast. Nature 38: 642–646. - PubMed
-
- Bessman, M.J., Frick, D.N, and O’Handley, S.F. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271: 25059–25062. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
