Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
- PMID: 12554651
- PMCID: PMC140724
- DOI: 10.1093/emboj/cdg039
Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
Abstract
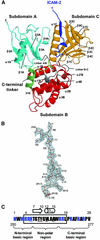
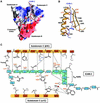
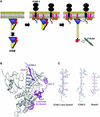
ERM (ezrin/radixin/moesin) proteins recognize the cytoplasmic domains of adhesion molecules in the formation of the membrane-associated cytoskeleton. Here we report the crystal structure of the radixin FERM (4.1 and ERM) domain complexed with the ICAM-2 cytoplasmic peptide. The non-polar region of the ICAM-2 peptide contains the RxxTYxVxxA sequence motif to form a beta-strand followed by a short 3(10)-helix. It binds the groove of the phosphotyrosine-binding (PTB)-like subdomain C mediated by a beta-beta association and several side-chain interactions. The binding mode of the ICAM-2 peptide to the FERM domain is distinct from that of the NPxY motif-containing peptide binding to the canonical PTB domain. Mutation analyses based on the crystal structure reveal the determinant elements of recognition and provide the first insights into the physical link between adhesion molecules and ERM proteins.
Figures



Similar articles
-
Structural basis for type II membrane protein binding by ERM proteins revealed by the radixin-neutral endopeptidase 24.11 (NEP) complex.J Biol Chem. 2007 Jul 6;282(27):19854-62. doi: 10.1074/jbc.M609232200. Epub 2007 Apr 24. J Biol Chem. 2007. PMID: 17459884
-
Crystallographic characterization of the radixin FERM domain bound to the cytoplasmic tail of the adhesion protein ICAM-2.Acta Crystallogr D Biol Crystallogr. 2001 Jun;57(Pt 6):891-2. doi: 10.1107/s0907444901005716. Epub 2001 May 25. Acta Crystallogr D Biol Crystallogr. 2001. PMID: 11375520
-
Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins.J Mol Biol. 2008 Sep 5;381(3):634-44. doi: 10.1016/j.jmb.2008.05.085. Epub 2008 Jun 7. J Mol Biol. 2008. PMID: 18614175
-
Radixin: cytoskeletal adopter and signaling protein.Int J Biochem Cell Biol. 2004 Nov;36(11):2131-6. doi: 10.1016/j.biocel.2003.11.018. Int J Biochem Cell Biol. 2004. PMID: 15313460 Review.
-
Two Sides of the Coin: Ezrin/Radixin/Moesin and Merlin Control Membrane Structure and Contact Inhibition.Int J Mol Sci. 2019 Apr 23;20(8):1996. doi: 10.3390/ijms20081996. Int J Mol Sci. 2019. PMID: 31018575 Free PMC article. Review.
Cited by
-
Ezrin interacts with the SARS coronavirus Spike protein and restrains infection at the entry stage.PLoS One. 2012;7(11):e49566. doi: 10.1371/journal.pone.0049566. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185364 Free PMC article.
-
Fosmid-based structure-function analysis reveals functionally distinct domains in the cytoplasmic domain of Drosophila crumbs.G3 (Bethesda). 2013 Feb;3(2):153-65. doi: 10.1534/g3.112.005074. Epub 2013 Feb 1. G3 (Bethesda). 2013. PMID: 23390593 Free PMC article.
-
A sequence conserved between CD5 and CD6 binds an FERM domain and exerts a restraint on T-cell activation.Immunology. 2019 Mar;156(3):270-276. doi: 10.1111/imm.13025. Epub 2018 Dec 10. Immunology. 2019. PMID: 30460991 Free PMC article.
-
Interactome analysis reveals ezrin can adopt multiple conformational states.J Biol Chem. 2013 Dec 6;288(49):35437-51. doi: 10.1074/jbc.M113.505669. Epub 2013 Oct 22. J Biol Chem. 2013. PMID: 24151071 Free PMC article.
-
ICAM-2 regulates vascular permeability and N-cadherin localization through ezrin-radixin-moesin (ERM) proteins and Rac-1 signalling.Cell Commun Signal. 2014 Mar 4;12:12. doi: 10.1186/1478-811X-12-12. Cell Commun Signal. 2014. PMID: 24593809 Free PMC article.
References
-
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. - PubMed
-
- Allenspach E.J. et al. (2001) ERM-dependent movement of CD43 defines a novel protein complex distal to the immunological synapse. Immunity, 15, 739–750. - PubMed
-
- Amieva M.R. and Furthmayr,H. (1995) Subcellular localization of moesin in dynamic filopodia, retraction fibers and other structures involved in substrate exploration, attachment and cell–cell contacts. Exp. Cell Res., 219, 180–196. - PubMed
-
- Andreoli C., Martin,M., Le Borgne,R., Reggio,H. and Mangeat,P. (1994) Ezrin has properties to self-associate at the plasma membrane. J. Cell Sci., 107, 2509–2521. - PubMed
-
- Arpin M., Algrain,M. and Louvard,D. (1994) Membrane–actin microfilament connections: an increasing diversity of players related to band 4.1. Curr. Opin. Cell Biol., 6, 136–141. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

