Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle
- PMID: 12529430
- PMCID: PMC140231
- DOI: 10.1091/mbc.e02-07-0376
Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle
Abstract
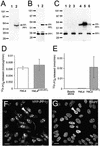
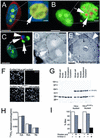
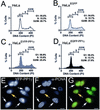
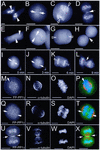
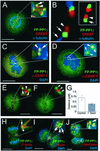
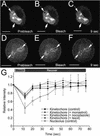
Protein phosphatase 1 (PP1) is a ubiquitous serine/threonine phosphatase that regulates many cellular processes, including cell division. When transiently expressed as fluorescent protein (FP) fusions, the three PP1 isoforms, alpha, beta/delta, and gamma1, are active phosphatases with distinct localization patterns. We report here the establishment and characterization of HeLa cell lines stably expressing either FP-PP1gamma or FP alone. Time-lapse imaging reveals dynamic targeting of FP-PP1gamma to specific sites throughout the cell cycle, contrasting with the diffuse pattern observed for FP alone. FP-PP1gamma shows a nucleolar accumulation during interphase. On entry into mitosis, it localizes initially at kinetochores, where it exchanges rapidly with the diffuse cytoplasmic pool. A dramatic relocalization of PP1 to the chromosome-containing regions occurs at the transition from early to late anaphase, and by telophase FP-PP1gamma also accumulates at the cleavage furrow and midbody. The changing spatio-temporal distribution of PP1gamma revealed using the stable PP1 cell lines implicates it in multiple processes, including nucleolar function, the regulation of chromosome segregation and cytokinesis.
Figures
Similar articles
-
Visualization of intracellular PP1 targeting through transiently and stably expressed fluorescent protein fusions.Methods Mol Biol. 2007;365:133-54. doi: 10.1385/1-59745-267-X:133. Methods Mol Biol. 2007. PMID: 17200560
-
NOM1 targets protein phosphatase I to the nucleolus.J Biol Chem. 2008 Jan 4;283(1):398-404. doi: 10.1074/jbc.M706708200. Epub 2007 Oct 26. J Biol Chem. 2008. PMID: 17965019
-
Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells.J Cell Sci. 2001 Dec;114(Pt 23):4219-28. doi: 10.1242/jcs.114.23.4219. J Cell Sci. 2001. PMID: 11739654
-
Protein phosphatases and cell division cycle control.Ciba Found Symp. 1992;170:130-40; discussion 140-6. doi: 10.1002/9780470514320.ch9. Ciba Found Symp. 1992. PMID: 1336448 Review.
-
Functional diversity of protein phosphatase-1, a cellular economizer and reset button.Physiol Rev. 2004 Jan;84(1):1-39. doi: 10.1152/physrev.00013.2003. Physiol Rev. 2004. PMID: 14715909 Review.
Cited by
-
New insights into the mechanism for chromosome alignment in metaphase.Int Rev Cell Mol Biol. 2013;303:237-62. doi: 10.1016/B978-0-12-407697-6.00006-4. Int Rev Cell Mol Biol. 2013. PMID: 23445812 Free PMC article. Review.
-
Connecting up and clearing out: how kinetochore attachment silences the spindle assembly checkpoint.Chromosoma. 2012 Oct;121(5):509-25. doi: 10.1007/s00412-012-0378-5. Epub 2012 Jul 11. Chromosoma. 2012. PMID: 22782189 Review.
-
Nuclear lamin phosphorylation: an emerging role in gene regulation and pathogenesis of laminopathies.Nucleus. 2020 Dec;11(1):299-314. doi: 10.1080/19491034.2020.1832734. Nucleus. 2020. PMID: 33030403 Free PMC article. Review.
-
Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins.Curr Biol. 2007 May 1;17(9):749-60. doi: 10.1016/j.cub.2007.03.064. Epub 2007 Apr 19. Curr Biol. 2007. PMID: 17446074 Free PMC article.
-
Cell division: control of the chromosomal passenger complex in time and space.Chromosoma. 2014 Mar;123(1-2):25-42. doi: 10.1007/s00412-013-0437-6. Epub 2013 Oct 3. Chromosoma. 2014. PMID: 24091645 Free PMC article. Review.
References
-
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. - PubMed
-
- Barker HM, Brewis ND, Street AJ, Spurr NK, Cohen PT. Three genes for protein phosphatase 1 map to different human chromosomes: sequence, expression and gene localization of protein serine/threonine phosphatase 1 beta (PPP1CB) Biochim Biophys Acta. 1994;1220:212–218. - PubMed
-
- Barker HM, Craig SP, Spurr NK, Cohen PT. Sequence of human protein serine/threonine phosphatase 1 gamma and localization of the gene (PPP1CC) encoding it to chromosome bands 12q24.1-q24.2. Biochim Biophys Acta. 1993;1178:228–233. - PubMed
-
- Berndt N, Dohadwala M, Liu CW. Constitutively active protein phosphatase 1alpha causes Rb-dependent G1 arrest in human cancer cells. Curr Biol. 1997;7:375–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

