ArrayExpress--a public repository for microarray gene expression data at the EBI
- PMID: 12519949
- PMCID: PMC165538
- DOI: 10.1093/nar/gkg091
ArrayExpress--a public repository for microarray gene expression data at the EBI
Abstract
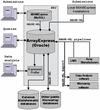
ArrayExpress is a new public database of microarray gene expression data at the EBI, which is a generic gene expression database designed to hold data from all microarray platforms. ArrayExpress uses the annotation standard Minimum Information About a Microarray Experiment (MIAME) and the associated XML data exchange format Microarray Gene Expression Markup Language (MAGE-ML) and it is designed to store well annotated data in a structured way. The ArrayExpress infrastructure consists of the database itself, data submissions in MAGE-ML format or via an online submission tool MIAMExpress, online database query interface, and the Expression Profiler online analysis tool. ArrayExpress accepts three types of submission, arrays, experiments and protocols, each of these is assigned an accession number. Help on data submission and annotation is provided by the curation team. The database can be queried on parameters such as author, laboratory, organism, experiment or array types. With an increasing number of organisations adopting MAGE-ML standard, the volume of submissions to ArrayExpress is increasing rapidly. The database can be accessed at http://www.ebi.ac.uk/arrayexpress.
Figures
Similar articles
-
ArrayExpress--a public repository for microarray gene expression data at the EBI.Nucleic Acids Res. 2005 Jan 1;33(Database issue):D553-5. doi: 10.1093/nar/gki056. Nucleic Acids Res. 2005. PMID: 15608260 Free PMC article.
-
ArrayExpress: a public database of gene expression data at EBI.C R Biol. 2003 Oct-Nov;326(10-11):1075-8. doi: 10.1016/j.crvi.2003.09.026. C R Biol. 2003. PMID: 14744115
-
Plant-based microarray data at the European Bioinformatics Institute. Introducing AtMIAMExpress, a submission tool for Arabidopsis gene expression data to ArrayExpress.Plant Physiol. 2005 Oct;139(2):632-6. doi: 10.1104/pp.105.063156. Plant Physiol. 2005. PMID: 16219923 Free PMC article.
-
Data storage and analysis in ArrayExpress.Methods Enzymol. 2006;411:370-86. doi: 10.1016/S0076-6879(06)11020-4. Methods Enzymol. 2006. PMID: 16939801 Review.
-
MGED standards: work in progress.OMICS. 2006 Summer;10(2):138-44. doi: 10.1089/omi.2006.10.138. OMICS. 2006. PMID: 16901218 Review.
Cited by
-
Overview of DNA microarrays: types, applications, and their future.Curr Protoc Mol Biol. 2013 Jan;Chapter 22:Unit 22.1.. doi: 10.1002/0471142727.mb2201s101. Curr Protoc Mol Biol. 2013. PMID: 23288464 Free PMC article.
-
Expression of microRNAs in the NCI-60 cancer cell-lines.PLoS One. 2012;7(11):e49918. doi: 10.1371/journal.pone.0049918. Epub 2012 Nov 28. PLoS One. 2012. PMID: 23209617 Free PMC article.
-
Identifying module biomarker in type 2 diabetes mellitus by discriminative area of functional activity.BMC Bioinformatics. 2015 Mar 18;16:92. doi: 10.1186/s12859-015-0519-y. BMC Bioinformatics. 2015. PMID: 25888350 Free PMC article.
-
PHIDIAS: a pathogen-host interaction data integration and analysis system.Genome Biol. 2007;8(7):R150. doi: 10.1186/gb-2007-8-7-r150. Genome Biol. 2007. PMID: 17663773 Free PMC article.
-
Practical application of toxicogenomics for profiling toxicant-induced biological perturbations.Int J Mol Sci. 2010 Sep 20;11(9):3397-412. doi: 10.3390/ijms11093397. Int J Mol Sci. 2010. PMID: 20957103 Free PMC article. Review.
References
-
- The Chipping Forecast (1999) Suppl. Nature Genet., 21, 1–60.
-
- Brazma A., Robinson,A., Cameron,G. and Ashburner,M. (2000) One-stop shop for microarray data. Nature, 403, 699–700. - PubMed
-
- Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C., Gaasterland,T., Glenisson,P., Holstege,F.C.P., Kim,I.F., Markowitz,V., Matese,J.C., Parkinson,H., Robinson,A., Sarkans,U., Schulze-Kremer,S., Stewart,J., Taylor,R., Vilo,J. and Vingron,M. (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genet., 29, 365–371. - PubMed
-
- Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M., Swiatek,M., Marks,W.L., Goncalves,J., Markel,S., Iordan,D., Shojatalab,M., Pizarro,A., White,J., Hubley,R., Deutsch,E., Senger,M., Aronow,B.J., Robinson,A., Bassett,D., Stoeckert,C.J.,Jr. and Brazma,A. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, research0046.1–0046.9. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources