Weak alignment offers new NMR opportunities to study protein structure and dynamics
- PMID: 12493823
- PMCID: PMC2312400
- DOI: 10.1110/ps.0233303
Weak alignment offers new NMR opportunities to study protein structure and dynamics
Abstract
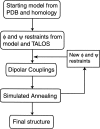
Protein solution nuclear magnetic resonance (NMR) can be conducted in a slightly anisotropic environment, where the orientational distribution of the proteins is no longer random. In such an environment, the large one-bond internuclear dipolar interactions no longer average to zero and report on the average orientation of the corresponding vectors relative to the magnetic field. The desired very weak ordering, on the order of 10(-3), can be induced conveniently by the use of aqueous nematic liquid crystalline suspensions or by anisotropically compressed hydrogels. The resulting residual dipolar interactions are scaled down by three orders of magnitude relative to their static values, but nevertheless can be measured at high accuracy. They are very precise reporters on the average orientation of bonds relative to the molecular alignment frame, and they can be used in a variety of ways to enrich our understanding of protein structure and function. Applications to date have focused primarily on validation of structures, determined by NMR, X-ray crystallography, or homology modeling, and on refinement of structures determined by conventional NMR approaches. Although de novo structure determination on the basis of dipolar couplings suffers from a severe multiple minimum problem, related to the degeneracy of dipolar coupling relative to inversion of the internuclear vector, a number of approaches can address this problem and potentially can accelerate the NMR structure determination process considerably. In favorable cases, where large numbers of dipolar couplings can be measured, inconsistency between measured values can report on internal motions.
Figures



Similar articles
-
Residual dipolar couplings in protein structure determination.Methods Mol Biol. 2004;278:89-106. doi: 10.1385/1-59259-809-9:089. Methods Mol Biol. 2004. PMID: 15317993
-
Weak alignment NMR: a hawk-eyed view of biomolecular structure.Curr Opin Struct Biol. 2005 Oct;15(5):563-70. doi: 10.1016/j.sbi.2005.08.006. Curr Opin Struct Biol. 2005. PMID: 16140525 Review.
-
Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy.J Am Chem Soc. 2003 Jul 30;125(30):9179-91. doi: 10.1021/ja0350684. J Am Chem Soc. 2003. PMID: 15369375
-
Orienting domains in proteins using dipolar couplings measured by liquid-state NMR: differences in solution and crystal forms of maltodextrin binding protein loaded with beta-cyclodextrin.J Mol Biol. 2000 Feb 4;295(5):1265-73. doi: 10.1006/jmbi.1999.3430. J Mol Biol. 2000. PMID: 10653702
-
Homonuclear dipolar recoupling techniques for structure determination in uniformly 13C-labeled proteins.Solid State Nucl Magn Reson. 2009 Nov;36(3):119-28. doi: 10.1016/j.ssnmr.2009.07.003. Epub 2009 Aug 5. Solid State Nucl Magn Reson. 2009. PMID: 19729285 Review.
Cited by
-
Deconvoluting Protein (Un)folding Structural Ensembles Using X-Ray Scattering, Nuclear Magnetic Resonance Spectroscopy and Molecular Dynamics Simulation.PLoS One. 2015 May 6;10(5):e0125662. doi: 10.1371/journal.pone.0125662. eCollection 2015. PLoS One. 2015. PMID: 25946337 Free PMC article.
-
Application of correlated residual dipolar couplings to the determination of the molecular alignment tensor magnitude of oriented proteins and nucleic acids.J Biomol NMR. 2004 Mar;28(3):273-87. doi: 10.1023/B:JNMR.0000013701.16162.0c. J Biomol NMR. 2004. PMID: 14752260
-
Structure of calmodulin complexed with an olfactory CNG channel fragment and role of the central linker: residual dipolar couplings to evaluate calmodulin binding modes outside the kinase family.J Biomol NMR. 2005 Mar;31(3):185-99. doi: 10.1007/s10858-005-0165-1. J Biomol NMR. 2005. PMID: 15803393
-
Structural assembly of molecular complexes based on residual dipolar couplings.J Am Chem Soc. 2010 Jul 7;132(26):8961-72. doi: 10.1021/ja100447p. J Am Chem Soc. 2010. PMID: 20550109 Free PMC article.
-
Structure and functional implications of a complex containing a segment of U6 RNA bound by a domain of Prp24.RNA. 2010 Apr;16(4):792-804. doi: 10.1261/rna.1913310. Epub 2010 Feb 24. RNA. 2010. PMID: 20181740 Free PMC article.
References
-
- Almond, A. and Axelsen, J.B. 2002. Physical interpretation of residual dipolar couplings in neutral aligned media. J. Am. Chem. Soc. 124 9986–9987. - PubMed
-
- Annila, A., Aitio, H., Thulin, E., and Drakenberg, T. 1999. Recognition of protein folds via dipolar couplings. J. Biomol. NMR 14 223–230.
-
- Banci, L., Bertini, I., Savellini, G.G., Romagnoli, A., Turano, P., Cremonini, M.A., Luchinat, C., and Gray, H.B. 1997. Pseudocontact shifts as constraints for energy minimization and molecular dynamics calculations on solution structures of paramagnetic metalloproteins. Proteins-Structure Function and Genetics 29 68–76. - PubMed
-
- Barbato, G., Ikura, M., Kay, L.E., Pastor, R.W., Bax, A. 1992. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry 31 5269–5278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

