Rotavirus gene silencing by small interfering RNAs
- PMID: 12446562
- PMCID: PMC1308328
- DOI: 10.1093/embo-reports/kvf234
Rotavirus gene silencing by small interfering RNAs
Abstract
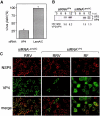
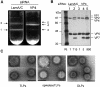
RNA interference is an evolutionarily conserved double-stranded RNA-triggered mechanism for suppressing gene expression. Rotaviruses, the leading cause of severe diarrhea in young children, are formed by three concentric layers of protein, from which the spike protein VP4 projects. Here, we show that a small interfering RNA corresponding to the VP4 gene efficiently inhibits the synthesis of this protein in virus-infected cells. A large proportion of infected cells had no detectable VP4 and the yield of viral progeny was reduced. Most of the virus particles purified from these cells were triple-layered, but lacked VP4, and were poorly infectious. We also show that VP4 might not be required for the last step of virus morphogenesis. The VP4 gene silencing was specific, since the synthesis of VP4 from rotavirus strains that differ in the target sequence was not affected. These findings offer the possibility of carrying out reverse genetics in rotaviruses.
Figures

Similar articles
-
Silencing the morphogenesis of rotavirus.J Virol. 2005 Jan;79(1):184-92. doi: 10.1128/JVI.79.1.184-192.2005. J Virol. 2005. PMID: 15596814 Free PMC article.
-
Actin-Dependent Nonlytic Rotavirus Exit and Infectious Virus Morphogenetic Pathway in Nonpolarized Cells.J Virol. 2018 Feb 26;92(6):e02076-17. doi: 10.1128/JVI.02076-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263265 Free PMC article.
-
The C Terminus of Rotavirus VP4 Protein Contains an Actin Binding Domain Which Requires Cooperation with the Coiled-Coil Domain for Actin Remodeling.J Virol. 2018 Dec 10;93(1):e01598-18. doi: 10.1128/JVI.01598-18. Print 2019 Jan 1. J Virol. 2018. PMID: 30333172 Free PMC article.
-
RNA silencing of rotavirus gene expression.Virus Res. 2004 Jun 1;102(1):43-51. doi: 10.1016/j.virusres.2004.01.014. Virus Res. 2004. PMID: 15068879 Free PMC article. Review.
-
Molecular biology of rotavirus cell entry.Arch Med Res. 2002 Jul-Aug;33(4):356-61. doi: 10.1016/s0188-4409(02)00374-0. Arch Med Res. 2002. PMID: 12234525 Review.
Cited by
-
Rotavirus and Serotonin Cross-Talk in Diarrhoea.PLoS One. 2016 Jul 26;11(7):e0159660. doi: 10.1371/journal.pone.0159660. eCollection 2016. PLoS One. 2016. PMID: 27459372 Free PMC article.
-
Inhibition of Tulane virus replication in vitro with RNA interference.J Med Virol. 2013 Jan;85(1):179-86. doi: 10.1002/jmv.23340. J Med Virol. 2013. PMID: 23154881 Free PMC article.
-
Rotavirus Nonstructural Protein NSP3 is not required for viral protein synthesis.J Virol. 2006 Sep;80(18):9031-8. doi: 10.1128/JVI.00437-06. J Virol. 2006. PMID: 16940515 Free PMC article.
-
The N- and C-terminal regions of rotavirus NSP5 are the critical determinants for the formation of viroplasm-like structures independent of NSP2.J Virol. 2003 Nov;77(22):12184-92. doi: 10.1128/jvi.77.22.12184-12192.2003. J Virol. 2003. PMID: 14581555 Free PMC article.
-
Rotavirus Spike Protein VP4 Mediates Viroplasm Assembly by Association to Actin Filaments.J Virol. 2022 Sep 14;96(17):e0107422. doi: 10.1128/jvi.01074-22. Epub 2022 Aug 8. J Virol. 2022. PMID: 35938869 Free PMC article.
References
-
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K. and Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. - PubMed
-
- Estes M.K. (1996) Rotaviruses and their replication. In Fields, B.N. et al. . (eds), Virology. Raven Press, New York, Vol. 2, pp. 1625–1655.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

