Control of biochemical reactions through supramolecular RING domain self-assembly
- PMID: 12438698
- PMCID: PMC137729
- DOI: 10.1073/pnas.202608799
Control of biochemical reactions through supramolecular RING domain self-assembly
Abstract
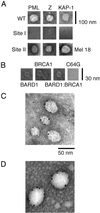
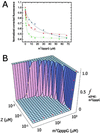
RING domains act in a variety of unrelated biochemical reactions, with many of these domains forming key parts of supramolecular assemblies in cells. Here, we observe that purified RINGs from a variety of functionally unrelated proteins, including promyelocytic leukemia protein, KAP-1TIF1beta, Z, Mel18, breast cancer susceptibility gene product 1 (BRCA1), and BRCA1-associated RING domain (BARD1), self-assemble into supramolecular structures in vitro that resemble those they form in cells. RING bodies form polyvalent binding surfaces and scaffold multiple partner proteins. Separation of RING bodies from monomers reveals that self-assembly controls and amplifies their specific activities in two unrelated biochemistries: reduction of 5' mRNA cap affinity of eIF4E by promyelocytic leukemia protein and Z, and E3 ubiquitin conjugation activity of BARD1:BRCA1. Functional significance of self-assembly is underscored by partial restoration of assembly and E3 activity of cancer predisposing BRCA1 mutant by forced oligomerization. RING self-assembly creates bodies that act structurally as polyvalent scaffolds, thermodynamically by amplifying activities of partner proteins, and catalytically by spatiotemporal coupling of enzymatic reactions. These studies reveal a general paradigm of how supramolecular structures may function in cells.
Figures


Similar articles
-
The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E.J Mol Biol. 2001 Sep 28;312(4):609-23. doi: 10.1006/jmbi.2001.5003. J Mol Biol. 2001. PMID: 11575918
-
Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex.Proc Natl Acad Sci U S A. 2003 May 13;100(10):5646-51. doi: 10.1073/pnas.0836054100. Epub 2003 May 5. Proc Natl Acad Sci U S A. 2003. PMID: 12732733 Free PMC article.
-
The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity.Curr Opin Genet Dev. 2002 Feb;12(1):86-91. doi: 10.1016/s0959-437x(01)00269-6. Curr Opin Genet Dev. 2002. PMID: 11790560 Review.
-
Structure of a BRCA1-BARD1 heterodimeric RING-RING complex.Nat Struct Biol. 2001 Oct;8(10):833-7. doi: 10.1038/nsb1001-833. Nat Struct Biol. 2001. PMID: 11573085
-
RING fingers and B-boxes: zinc-binding protein-protein interaction domains.Biochem Cell Biol. 1998;76(2-3):351-8. doi: 10.1139/bcb-76-2-3-351. Biochem Cell Biol. 1998. PMID: 9923704 Review.
Cited by
-
TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6.Proc Natl Acad Sci U S A. 2004 Oct 26;101(43):15318-23. doi: 10.1073/pnas.0404132101. Epub 2004 Oct 18. Proc Natl Acad Sci U S A. 2004. PMID: 15492226 Free PMC article.
-
A RING domain gene is expressed in different cell types of leaf trace, stem, and juvenile bundles in the stem vascular system of zinnia.Plant Physiol. 2005 Jul;138(3):1383-95. doi: 10.1104/pp.104.057901. Epub 2005 Jun 17. Plant Physiol. 2005. PMID: 15965022 Free PMC article.
-
Mechanism of p53 stabilization by ATM after DNA damage.Cell Cycle. 2010 Feb 1;9(3):472-8. doi: 10.4161/cc.9.3.10556. Cell Cycle. 2010. PMID: 20081365 Free PMC article.
-
Parkin, A Top Level Manager in the Cell's Sanitation Department.Open Biochem J. 2011;5:9-26. doi: 10.2174/1874091X01105010009. Epub 2011 Apr 18. Open Biochem J. 2011. PMID: 21633666 Free PMC article.
-
Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates.Cancers (Basel). 2022 Sep 20;14(19):4549. doi: 10.3390/cancers14194549. Cancers (Basel). 2022. PMID: 36230470 Free PMC article. Review.
References
-
- Melnick A. & Licht, J. D. (1999) Blood 93 3167-3215. - PubMed
-
- Chen Y., Chen, C. F., Riley, D. J., Allred, D. C., Chen, P. L., Von Hoff, D., Osborne, C. K. & Lee, W. H. (1995) Science 270 789-791. - PubMed
-
- Kentsis A. & Borden, K. L. B. (2000) Curr. Protein Pept. Sci. 1 49-73. - PubMed
-
- Saurin A. J., Borden, K. L., Boddy, M. N. & Freemont, P. S. (1996) Trends Biochem. Sci. 21 208-214. - PubMed
-
- Peng H., Begg, G. E., Schultz, D. C., Friedman, J. R., Jensen, D. E., Speicher, D. W. & Rauscher, F. J., III (2000) J. Mol. Biol. 295 1139-1162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

