Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands
- PMID: 12429062
- PMCID: PMC133447
- DOI: 10.1186/gb-2002-3-11-research0063
Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands
Abstract
Background: G-protein-coupled receptors (GPCRs) are the largest and most diverse family of transmembrane receptors. They respond to a wide range of stimuli, including small peptides, lipid analogs, amino-acid derivatives, and sensory stimuli such as light, taste and odor, and transmit signals to the interior of the cell through interaction with heterotrimeric G proteins. A large number of putative GPCRs have no identified natural ligand. We hypothesized that a more complete knowledge of the phylogenetic relationship of these orphan receptors to receptors with known ligands could facilitate ligand identification, as related receptors often have ligands with similar structural features.
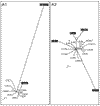
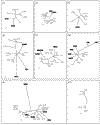
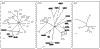
Results: A database search excluding olfactory and gustatory receptors was used to compile a list of accession numbers and synonyms of 81 orphan and 196 human GPCRs with known ligands. Of these, 241 sequences belonging to the rhodopsin receptor-like family A were aligned and a tentative phylogenetic tree constructed by neighbor joining. This tree and local alignment tools were used to define 19 subgroups of family A small enough for more accurate maximum-likelihood analyses. The secretin receptor-like family B and metabotropic glutamate receptor-like family C were directly subjected to these methods.
Conclusions: Our trees show the overall relationship of 277 GPCRs with emphasis on orphan receptors. Support values are given for each branch. This approach may prove valuable for identification of the natural ligands of orphan receptors as their relation to receptors with known ligands becomes more evident.
Figures




Similar articles
-
Cross genome phylogenetic analysis of human and Drosophila G protein-coupled receptors: application to functional annotation of orphan receptors.BMC Genomics. 2005 Aug 10;6:106. doi: 10.1186/1471-2164-6-106. BMC Genomics. 2005. PMID: 16091152 Free PMC article.
-
Deriving structural and functional insights from a ligand-based hierarchical classification of G protein-coupled receptors.Protein Eng. 2002 Jan;15(1):7-12. doi: 10.1093/protein/15.1.7. Protein Eng. 2002. PMID: 11842232
-
Family-B G-protein-coupled receptors.Genome Biol. 2001;2(12):REVIEWS3013. doi: 10.1186/gb-2001-2-12-reviews3013. Epub 2001 Nov 23. Genome Biol. 2001. PMID: 11790261 Free PMC article. Review.
-
An expressed sequence tag (EST) data mining strategy succeeding in the discovery of new G-protein coupled receptors.J Mol Biol. 2001 Mar 30;307(3):799-813. doi: 10.1006/jmbi.2001.4520. J Mol Biol. 2001. PMID: 11273702
-
G-Protein coupled receptors: models, mutagenesis, and drug design.J Med Chem. 1998 Jul 30;41(16):2911-27. doi: 10.1021/jm970767a. J Med Chem. 1998. PMID: 9685229 Review. No abstract available.
Cited by
-
Protein 3D structure computed from evolutionary sequence variation.PLoS One. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. Epub 2011 Dec 7. PLoS One. 2011. PMID: 22163331 Free PMC article.
-
Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin.Biochemistry. 2003 Mar 18;42(10):2759-67. doi: 10.1021/bi027224+. Biochemistry. 2003. PMID: 12627940 Free PMC article. Review.
-
Anatomical profiling of G protein-coupled receptor expression.Cell. 2008 Oct 31;135(3):561-71. doi: 10.1016/j.cell.2008.08.040. Cell. 2008. PMID: 18984166 Free PMC article.
-
CD4+ T-cell subsets in inflammatory diseases: beyond the Th1/Th2 paradigm.Int Immunol. 2016 Apr;28(4):163-71. doi: 10.1093/intimm/dxw006. Epub 2016 Feb 12. Int Immunol. 2016. PMID: 26874355 Free PMC article. Review.
-
Hunting for the function of orphan GPCRs - beyond the search for the endogenous ligand.Br J Pharmacol. 2015 Jul;172(13):3212-28. doi: 10.1111/bph.12942. Epub 2014 Dec 15. Br J Pharmacol. 2015. PMID: 25231237 Free PMC article. Review.
References
-
- Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. - PubMed
-
- Libert F, Parmentier M, Lefort A, Dinsart C, Van Sande J, Maenhaut C, Simons MJ, Dumont JE, Vassart G. Selective amplification and cloning of four new members of the G protein-coupled receptor family. Science. 1989;244:569–572. - PubMed
-
- Methner A, Hermey G, Schinke B, Hermans-Borgmeyer I. A novel G protein-coupled receptor with homology to neuropeptide and chemoattractant receptors expressed during bone development. Biochem Biophys Res Commun. 1997;233:336–342. - PubMed
-
- Lee D, George S, Evans J, Lynch K, O'Dowd B. Orphan G protein-coupled receptors in the CNS. Curr Opin Pharmacol. 2001;1:31–39. - PubMed
-
- Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR, Grandy DK, Langen H, Monsma FJ, Jr, Civelli O. Orphanin FQ: a neuropeptide that activates an opioidlike G protein-coupled receptor. Science. 1995;270:792–794. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

