Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
- PMID: 12426397
- PMCID: PMC137216
- DOI: 10.1093/emboj/cdf627
Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
Abstract
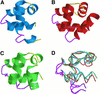
The C-terminal domain of the UvrC protein (UvrC CTD) is essential for 5' incision in the prokaryotic nucleotide excision repair process. We have determined the three-dimensional structure of the UvrC CTD using heteronuclear NMR techniques. The structure shows two helix-hairpin-helix (HhH) motifs connected by a small connector helix. The UvrC CTD is shown to mediate structure-specific DNA binding. The domain binds to a single-stranded-double-stranded junction DNA, with a strong specificity towards looped duplex DNA that contains at least six unpaired bases per loop ("bubble DNA"). Using chemical shift perturbation experiments, the DNA-binding surface is mapped to the first hairpin region encompassing the conserved glycine-valine-glycine residues followed by lysine-arginine-arginine, a positively charged surface patch and the second hairpin region consisting of glycine-isoleucine-serine. A model for the protein-DNA complex is proposed that accounts for this specificity.
Figures

Similar articles
-
The C-terminal region of the Escherichia coli UvrC protein, which is homologous to the C-terminal region of the human ERCC1 protein, is involved in DNA binding and 5'-incision.Nucleic Acids Res. 1998 Jan 15;26(2):462-8. doi: 10.1093/nar/26.2.462. Nucleic Acids Res. 1998. PMID: 9421501 Free PMC article.
-
The C-terminal region of Escherichia coli UvrC contributes to the flexibility of the UvrABC nucleotide excision repair system.Nucleic Acids Res. 2002 Jun 1;30(11):2492-500. doi: 10.1093/nar/30.11.2492. Nucleic Acids Res. 2002. PMID: 12034838 Free PMC article.
-
Three-dimensional structural views of damaged-DNA recognition: T4 endonuclease V, E. coli Vsr protein, and human nucleotide excision repair factor XPA.Mutat Res. 2000 Aug 30;460(3-4):257-75. doi: 10.1016/s0921-8777(00)00031-8. Mutat Res. 2000. PMID: 10946233 Review.
-
Crystal structure of Escherichia coli UvrB C-terminal domain, and a model for UvrB-uvrC interaction.FEBS Lett. 2000 Jan 14;465(2-3):161-4. doi: 10.1016/s0014-5793(99)01690-7. FEBS Lett. 2000. PMID: 10631326
-
Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair.Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1467-72. doi: 10.1073/pnas.032584099. Epub 2002 Jan 29. Proc Natl Acad Sci U S A. 2002. PMID: 11818552 Free PMC article.
Cited by
-
Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes.Nucleic Acids Res. 2013 May;41(9):4901-12. doi: 10.1093/nar/gkt177. Epub 2013 Mar 19. Nucleic Acids Res. 2013. PMID: 23511970 Free PMC article.
-
Domain organization of DNase from Thioalkalivibrio sp. provides insights into retention of activity in high salt environments.Front Microbiol. 2015 Jul 1;6:661. doi: 10.3389/fmicb.2015.00661. eCollection 2015. Front Microbiol. 2015. PMID: 26191053 Free PMC article.
-
PCNA and XPF cooperate to distort DNA substrates.Nucleic Acids Res. 2010 Mar;38(5):1664-75. doi: 10.1093/nar/gkp1104. Epub 2009 Dec 11. Nucleic Acids Res. 2010. PMID: 20008103 Free PMC article.
-
Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding.Mol Cell. 2008 Jan 18;29(1):122-33. doi: 10.1016/j.molcel.2007.10.026. Epub 2007 Dec 27. Mol Cell. 2008. PMID: 18158267 Free PMC article.
-
DNA repair gets physical: mapping an XPA-binding site on ERCC1.DNA Repair (Amst). 2008 May 3;7(5):819-26. doi: 10.1016/j.dnarep.2008.01.018. Epub 2008 Mar 14. DNA Repair (Amst). 2008. PMID: 18343204 Free PMC article. Review.
References
-
- Bonvin A.M.J.J., Houben,K., Guenneugues,M., Kaptein,R. and Boelens,R. (2001) Rapid protein fold determination using secondary chemical shifts and cross-hydrogen bond 15N–13C scalar couplings (3hbJNC′). J. Biomol. NMR, 21, 221–233. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography a NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Cavanagh J., Fairbrother, WJ., Palmer,A.G.,III and Skelton,N.J. (1996) Protein NMR Spectroscopy. Academic Press, San Diego, CA.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

