MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation
- PMID: 12426395
- PMCID: PMC137207
- DOI: 10.1093/emboj/cdf616
MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation
Abstract
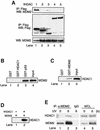
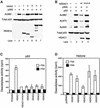
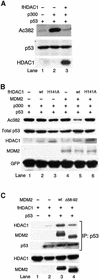
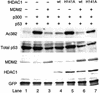
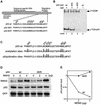
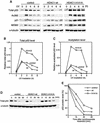
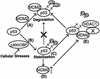
The tumor suppressor p53 is stabilized and activated in response to cellular stress through post-translational modifications including acetylation. p300/CBP-mediated acetylation of p53 is negatively regulated by MDM2. Here we show that MDM2 can promote p53 deacetylation by recruiting a complex containing HDAC1. The HDAC1 complex binds MDM2 in a p53-independent manner and deacetylates p53 at all known acetylated lysines in vivo. Ectopic expression of a dominant-negative HDAC1 mutant restores p53 acetylation in the presence of MDM2, whereas wild-type HDAC1 and MDM2 deacetylate p53 synergistically. Fibroblasts overexpressing a dominant negative HDAC1 mutant display enhanced DNA damage-induced p53 acetylation, increased levels of p53 and a more pronounced induction of p21 and MDM2. These results indicate that acetylation promotes p53 stability and function. As the acetylated p53 lysine residues overlap with those that are ubiquitylated, our results suggest that one major function of p53 acetylation is to promote p53 stability by preventing MDM2-dependent ubiquitylation, while recruitment of HDAC1 by MDM2 promotes p53 degradation by removing these acetyl groups.
Figures
Similar articles
-
MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins.Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12547-52. doi: 10.1073/pnas.97.23.12547. Proc Natl Acad Sci U S A. 2000. PMID: 11070080 Free PMC article.
-
Defective p53 post-translational modification required for wild type p53 inactivation in malignant epithelial cells with mdm2 gene amplification.J Biol Chem. 2003 Dec 26;278(52):52890-900. doi: 10.1074/jbc.M300279200. Epub 2003 Oct 10. J Biol Chem. 2003. PMID: 14555661
-
Acidic domain is indispensable for MDM2 to negatively regulate the acetylation of p53.Biochem Biophys Res Commun. 2008 Sep 26;374(3):437-41. doi: 10.1016/j.bbrc.2008.07.038. Epub 2008 Jul 16. Biochem Biophys Res Commun. 2008. PMID: 18638452
-
Dynamics of the p53 acetylation pathway.Novartis Found Symp. 2004;259:197-205; discussion 205-7, 223-5. Novartis Found Symp. 2004. PMID: 15171255 Review.
-
Regulation of p53: a collaboration between Mdm2 and Mdmx.Oncotarget. 2012 Mar;3(3):228-35. doi: 10.18632/oncotarget.443. Oncotarget. 2012. PMID: 22410433 Free PMC article. Review.
Cited by
-
It Takes Two to Tango: The Interplay between Prostate Cancer and Its Microenvironment from an Epigenetic Perspective.Cancers (Basel). 2024 Jan 10;16(2):294. doi: 10.3390/cancers16020294. Cancers (Basel). 2024. PMID: 38254784 Free PMC article. Review.
-
Extracellular vesicle-associated DNA: ten years since its discovery in human blood.Cell Death Dis. 2024 Sep 12;15(9):668. doi: 10.1038/s41419-024-07003-y. Cell Death Dis. 2024. PMID: 39266560 Free PMC article. Review.
-
Inauhzin and Nutlin3 synergistically activate p53 and suppress tumor growth.Cancer Biol Ther. 2012 Aug;13(10):915-24. doi: 10.4161/cbt.20844. Epub 2012 Aug 1. Cancer Biol Ther. 2012. PMID: 22785205 Free PMC article.
-
Dual Roles of MDM2 in the Regulation of p53: Ubiquitination Dependent and Ubiquitination Independent Mechanisms of MDM2 Repression of p53 Activity.Genes Cancer. 2012 Mar;3(3-4):240-8. doi: 10.1177/1947601912455199. Genes Cancer. 2012. PMID: 23150757 Free PMC article.
-
Mechanism, Consequences, and Therapeutic Targeting of Abnormal IL15 Signaling in Cutaneous T-cell Lymphoma.Cancer Discov. 2016 Sep;6(9):986-1005. doi: 10.1158/2159-8290.CD-15-1297. Epub 2016 Jul 15. Cancer Discov. 2016. PMID: 27422033 Free PMC article.
References
-
- Appella E. and Anderson,C.W. (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem., 268, 2764–2772. - PubMed
-
- Barlev N.A., Liu,L., Chehab,N.H., Mansfield,K., Harris,K.G., Halazonetis,T.D. and Berger,S.L. (2001) Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell, 8, 1243–1254. - PubMed
-
- Boyd S.D., Tsai,K.Y. and Jacks,T. (2000) An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol., 2, 563–568. - PubMed
-
- Canman C.E., Lim,D.S., Cimprich,K.A., Taya,Y., Tamai,K., Sakaguchi,K., Appella,E., Kastan,M.B. and Siliciano,J.D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science, 281, 1677–1679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

