Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p
- PMID: 12417727
- PMCID: PMC134069
- DOI: 10.1128/MCB.22.23.8241-8253.2002
Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p
Abstract
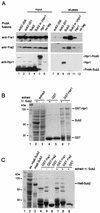
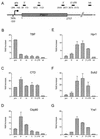
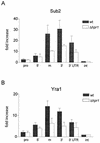
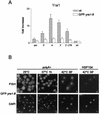
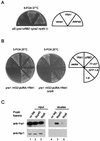
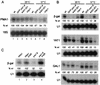
Yra1p/REF participates in mRNA export by recruiting the export receptor Mex67p to messenger ribonucleoprotein (mRNP) complexes. Yra1p also binds Sub2p, a DEAD box ATPase/RNA helicase implicated in splicing and required for mRNA export. We identified genetic and physical interactions between Yra1p, Sub2p, and Hpr1p, a protein involved in transcription elongation whose deletion leads to poly(A)(+) RNA accumulation in the nucleus. By chromatin immunoprecipitation (ChIP) experiments, we show that Hpr1p, Sub2p, and Yra1p become associated with active genes during transcription elongation and that Hpr1p is required for the efficient recruitment of Sub2p and Yra1p. The data indicate that transcription and export are functionally linked and that mRNA export defects may be due in part to inefficient loading of essential mRNA export factors on the growing mRNP. We also identified functional interactions between Yra1p and the exosome components Rrp45p and Rrp6p. We show that yra1, sub2, and Deltahpr1 mutants all present defects in mRNA accumulation and that deletion of RRP6 in yra1 mutants restores normal mRNA levels. The data support the hypothesis that an exosome-dependent surveillance mechanism targets improperly assembled mRNPs for degradation.
Figures

Similar articles
-
Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p.Nature. 2001 Oct 11;413(6856):648-52. doi: 10.1038/35098113. Nature. 2001. PMID: 11675790
-
Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes.EMBO J. 2004 Jul 7;23(13):2620-31. doi: 10.1038/sj.emboj.7600261. Epub 2004 Jun 10. EMBO J. 2004. PMID: 15192704 Free PMC article.
-
The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p.Mol Cell Biol. 2001 Jul;21(13):4219-32. doi: 10.1128/MCB.21.13.4219-4232.2001. Mol Cell Biol. 2001. PMID: 11390651 Free PMC article.
-
Nuclear export of mRNA.FEBS Lett. 2001 Jun 8;498(2-3):150-6. doi: 10.1016/s0014-5793(01)02482-6. FEBS Lett. 2001. PMID: 11412847 Review.
-
Nuclear export of messenger RNA.Results Probl Cell Differ. 2002;35:133-50. doi: 10.1007/978-3-540-44603-3_7. Results Probl Cell Differ. 2002. PMID: 11791404 Review. No abstract available.
Cited by
-
Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA.Nucleic Acids Res. 2013 Jan;41(2):1294-306. doi: 10.1093/nar/gks1188. Epub 2012 Dec 7. Nucleic Acids Res. 2013. PMID: 23222130 Free PMC article.
-
Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription.Proc Natl Acad Sci U S A. 2006 Oct 31;103(44):16376-81. doi: 10.1073/pnas.0607941103. Epub 2006 Oct 20. Proc Natl Acad Sci U S A. 2006. PMID: 17056718 Free PMC article.
-
The CCR4-NOT complex physically and functionally interacts with TRAMP and the nuclear exosome.PLoS One. 2009 Aug 25;4(8):e6760. doi: 10.1371/journal.pone.0006760. PLoS One. 2009. PMID: 19707589 Free PMC article.
-
Mud2 functions in transcription by recruiting the Prp19 and TREX complexes to transcribed genes.Nucleic Acids Res. 2018 Oct 12;46(18):9749-9763. doi: 10.1093/nar/gky640. Nucleic Acids Res. 2018. PMID: 30053068 Free PMC article.
-
3'-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation.EMBO J. 2006 Sep 20;25(18):4253-62. doi: 10.1038/sj.emboj.7601305. Epub 2006 Aug 31. EMBO J. 2006. PMID: 16946703 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
