Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation
- PMID: 12417714
- PMCID: PMC134064
- DOI: 10.1128/MCB.22.23.8101-8113.2002
Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation
Abstract
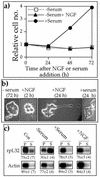
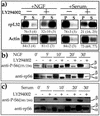
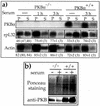
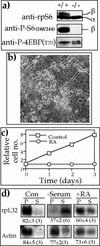
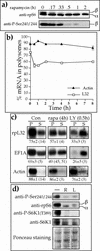
Translation of terminal oligopyrimidine tract (TOP) mRNAs, which encode multiple components of the protein synthesis machinery, is known to be controlled by mitogenic stimuli. We now show that the ability of cells to progress through the cell cycle is not a prerequisite for this mode of regulation. TOP mRNAs can be translationally activated when PC12 or embryonic stem (ES) cells are induced to grow (increase their size) by nerve growth factor and retinoic acid, respectively, while remaining mitotically arrested. However, both growth and mitogenic signals converge via the phosphatidylinositol 3-kinase (PI3-kinase)-mediated pathway and are transduced to efficiently translate TOP mRNAs. Translational activation of TOP mRNAs can be abolished by LY294002, a PI3-kinase inhibitor, or by overexpression of PTEN as well as by dominant-negative mutants of PI3-kinase or its effectors, PDK1 and protein kinase Balpha (PKBalpha). Likewise, overexpression of constitutively active PI3-kinase or PKBalpha can relieve the translational repression of TOP mRNAs in quiescent cells. Both mitogenic and growth signals lead to phosphorylation of ribosomal protein S6 (rpS6), which precedes the translational activation of TOP mRNAs. Nevertheless, neither rpS6 phosphorylation nor its kinase, S6K1, is essential for the translational response of these mRNAs. Thus, TOP mRNAs can be translationally activated by growth or mitogenic stimuli of ES cells, whose rpS6 is constitutively unphosphorylated due to the disruption of both alleles of S6K1. Similarly, complete inhibition of mammalian target of rapamycin (mTOR) and its effector S6K by rapamycin in various cell lines has only a mild repressive effect on the translation of TOP mRNAs. It therefore appears that translation of TOP mRNAs is primarily regulated by growth and mitogenic cues through the PI3-kinase pathway, with a minor role, if any, for the mTOR pathway.
Figures



Similar articles
-
S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway.Mol Cell Biol. 2004 Apr;24(8):3112-24. doi: 10.1128/MCB.24.8.3112-3124.2004. Mol Cell Biol. 2004. PMID: 15060135 Free PMC article.
-
Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation.Mol Cell Biol. 2001 Dec;21(24):8671-83. doi: 10.1128/MCB.21.24.8671-8683.2001. Mol Cell Biol. 2001. PMID: 11713299 Free PMC article.
-
Regulation of the phosphatidylinositol 3-kinase, Akt/protein kinase B, FRAP/mammalian target of rapamycin, and ribosomal S6 kinase 1 signaling pathways by thyroid-stimulating hormone (TSH) and stimulating type TSH receptor antibodies in the thyroid gland.J Biol Chem. 2003 Jun 13;278(24):21960-71. doi: 10.1074/jbc.M300805200. Epub 2003 Mar 30. J Biol Chem. 2003. PMID: 12668683
-
Ribosomal protein S6 kinase from TOP mRNAs to cell size.Prog Mol Biol Transl Sci. 2009;90:109-53. doi: 10.1016/S1877-1173(09)90003-5. Epub 2009 Oct 27. Prog Mol Biol Transl Sci. 2009. PMID: 20374740 Review.
-
Ribosomal protein S6 phosphorylation: from protein synthesis to cell size.Trends Biochem Sci. 2006 Jun;31(6):342-8. doi: 10.1016/j.tibs.2006.04.003. Epub 2006 May 6. Trends Biochem Sci. 2006. PMID: 16679021 Review.
Cited by
-
PKA-dependent phosphorylation of ribosomal protein S6 does not correlate with translation efficiency in striatonigral and striatopallidal medium-sized spiny neurons.J Neurosci. 2015 Mar 11;35(10):4113-30. doi: 10.1523/JNEUROSCI.3288-14.2015. J Neurosci. 2015. PMID: 25762659 Free PMC article.
-
S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway.Mol Cell Biol. 2004 Apr;24(8):3112-24. doi: 10.1128/MCB.24.8.3112-3124.2004. Mol Cell Biol. 2004. PMID: 15060135 Free PMC article.
-
Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability.Biochem J. 2003 Jun 1;372(Pt 2):555-66. doi: 10.1042/BJ20021266. Biochem J. 2003. PMID: 12611592 Free PMC article.
-
Rapamycin selectively reduces the association of transcripts containing complex 5' UTRs with ribosomes in C4-2B prostate cancer cells.J Cell Biochem. 2009 Jun 1;107(3):473-81. doi: 10.1002/jcb.22145. J Cell Biochem. 2009. PMID: 19347904 Free PMC article.
-
mTOR/S6 kinase pathway contributes to astrocyte survival during ischemia.J Biol Chem. 2009 Aug 14;284(33):22067-22078. doi: 10.1074/jbc.M109.033100. Epub 2009 Jun 17. J Biol Chem. 2009. PMID: 19535330 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
