RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes
- PMID: 12414928
- PMCID: PMC136867
- DOI: 10.1128/jvi.76.23.11853-11865.2002
RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes
Abstract
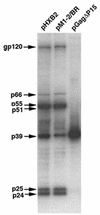
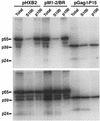
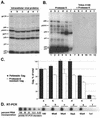
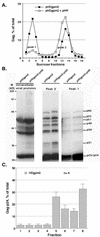
The nucleocapsid (NC) domain of retroviruses plays a critical role in specific viral RNA packaging and virus assembly. RNA is thought to facilitate viral particle assembly, but the results described here with NC mutants indicate that it also plays a critical role in particle integrity. We investigated the assembly and integrity of particles produced by the human immunodeficiency virus type 1 M1-2/BR mutant virus, in which 10 of the 13 positive residues of NC have been replaced with alanines and incorporation of viral genomic RNA is virtually abolished. We found that the mutations in the basic residues of NC did not disrupt Gag assembly at the cell membrane. The mutant Gag protein can assemble efficiently at the cell membrane, and viral proteins are detected outside the cell as efficiently as they are for the wild type. However, only approximately 10% of the Gag molecules present in the supernatant of this mutant sediment at the correct density for a retroviral particle. The reduction of positive charge in the NC basic domain of the M1-2/BR virus adversely affects both the specific and nonspecific RNA binding properties of NC, and thus the assembled Gag polyprotein does not bind significant amounts of viral or cellular RNA. We found a direct correlation between the percentage of Gag associated with sedimented particles and the amount of incorporated RNA. We conclude that RNA binding by Gag, whether the RNA is viral or not, is critical to retroviral particle integrity after cell membrane assembly and is less important for Gag-Gag interactions during particle assembly and release.
Figures

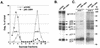


Similar articles
-
Mutations in the Basic Region of the Mason-Pfizer Monkey Virus Nucleocapsid Protein Affect Reverse Transcription, Genomic RNA Packaging, and the Virus Assembly Site.J Virol. 2018 Apr 27;92(10):e00106-18. doi: 10.1128/JVI.00106-18. Print 2018 May 15. J Virol. 2018. PMID: 29491167 Free PMC article.
-
Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA.J Virol. 2000 Apr;74(7):3046-57. doi: 10.1128/jvi.74.7.3046-3057.2000. J Virol. 2000. PMID: 10708419 Free PMC article.
-
Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses.J Virol. 2004 Jan;78(2):716-23. doi: 10.1128/jvi.78.2.716-723.2004. J Virol. 2004. PMID: 14694103 Free PMC article.
-
Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly.Semin Cell Dev Biol. 2019 Feb;86:129-139. doi: 10.1016/j.semcdb.2018.03.015. Epub 2018 Apr 1. Semin Cell Dev Biol. 2019. PMID: 29580971 Free PMC article. Review.
-
Properties and functions of the nucleocapsid protein in virus assembly.RNA Biol. 2010 Nov-Dec;7(6):744-53. doi: 10.4161/rna.7.6.14065. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21157181 Free PMC article. Review.
Cited by
-
Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably?Retrovirology. 2004 Sep 2;1:23. doi: 10.1186/1742-4690-1-23. Retrovirology. 2004. PMID: 15345057 Free PMC article. Review.
-
7SL RNA is retained in HIV-1 minimal virus-like particles as an S-domain fragment.J Virol. 2010 Sep;84(18):9070-7. doi: 10.1128/JVI.00714-10. Epub 2010 Jul 7. J Virol. 2010. PMID: 20610725 Free PMC article.
-
Functional analysis of the complex trans-activating response element RNA structure in simian immunodeficiency virus.J Virol. 2008 Sep;82(18):9171-8. doi: 10.1128/JVI.00530-08. Epub 2008 Jul 2. J Virol. 2008. PMID: 18596090 Free PMC article.
-
Detection of HIV-1 p24 Gag in plasma by a nanoparticle-based bio-barcode-amplification method.Nanomedicine (Lond). 2008 Jun;3(3):293-303. doi: 10.2217/17435889.3.3.293. Nanomedicine (Lond). 2008. PMID: 18510425 Free PMC article.
-
Identification of a minimal region of the HIV-1 5'-leader required for RNA dimerization, NC binding, and packaging.J Mol Biol. 2012 Mar 30;417(3):224-39. doi: 10.1016/j.jmb.2012.01.033. Epub 2012 Jan 27. J Mol Biol. 2012. PMID: 22306406 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

