The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor
- PMID: 12379151
- PMCID: PMC137586
- DOI: 10.1186/1471-2202-3-15
The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor
Abstract
Background: Huntington's disease (HD) pathogenesis is due to an expanded polyglutamine tract in huntingtin, but the specificity of neuronal loss compared with other polyglutamine disorders also implies a role for the protein's unknown inherent function. Huntingtin is moderately conserved, with 10 HEAT repeats reported in its amino-terminal half. HD orthologues are evident in vertebrates and Drosophila, but not in Saccharomyces cerevisiae, Caenorhabditis elegans or Arabidopsis thaliana, a phylogenetic profile similar to the NF-kB/Rel/dorsal family transcription factors, suggesting a potential functional relationship.
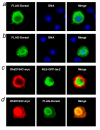
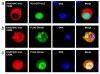
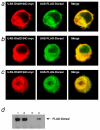
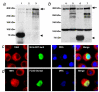
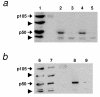
Results: We initially tested the potential for a relationship between huntingtin and dorsal by overexpression experiments in Drosophila S2 cells. Drosophila huntingtin complexes via its carboxyl-terminal region with dorsal, and the two enter the nucleus concomitantly, partly in a lipopolysaccharide (LPS)- and Nup88-dependent manner. Similarly, in HeLa cell extracts, human huntingtin co-immunoprecipitates with NF-kB p50 but not with p105. By cross-species comparative analysis, we find that the carboxyl-terminal segment of huntingtin that mediates the association with dorsal possesses numerous HEAT-like sequences related to those in the amino-terminal segment. Thus, Drosophila and vertebrate huntingtins are composed predominantly of 28 to 36 degenerate HEAT-like repeats that span the entire protein.
Conclusion: Like other HEAT-repeat filled proteins, huntingtin is made up largely of degenerate HEAT-like sequences, suggesting that it may play a scaffolding role in the formation of particular protein-protein complexes. While many proteins have been implicated in complexes with the amino-terminal region of huntingtin, the NF-kB/Rel/dorsal family transcription factors merit further examination as direct or indirect interactors with huntingtin's carboxyl-terminal segment.
Figures
Similar articles
-
Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture.Hum Mol Genet. 1998 May;7(5):783-90. doi: 10.1093/hmg/7.5.783. Hum Mol Genet. 1998. PMID: 9536081
-
Aggregation of N-terminal huntingtin is dependent on the length of its glutamine repeats.Hum Mol Genet. 1998 May;7(5):777-82. doi: 10.1093/hmg/7.5.777. Hum Mol Genet. 1998. PMID: 9536080
-
Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins.Somat Cell Mol Genet. 1998 Jul;24(4):217-33. doi: 10.1023/b:scam.0000007124.19463.e5. Somat Cell Mol Genet. 1998. PMID: 10410676
-
Targeting aggregation in the development of therapeutics for the treatment of Huntington's disease and other polyglutamine repeat diseases.Expert Opin Ther Targets. 2003 Apr;7(2):201-13. doi: 10.1517/14728222.7.2.201. Expert Opin Ther Targets. 2003. PMID: 12667098 Review.
-
Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease.Prog Neurobiol. 2007 Nov;83(4):211-27. doi: 10.1016/j.pneurobio.2006.11.004. Epub 2007 Jan 22. Prog Neurobiol. 2007. PMID: 17240517 Review.
Cited by
-
Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner.J Neurosci. 2008 Oct 15;28(42):10720-33. doi: 10.1523/JNEUROSCI.2126-08.2008. J Neurosci. 2008. PMID: 18923047 Free PMC article.
-
Is Huntingtin Dispensable in the Adult Brain?J Huntingtons Dis. 2017;6(1):1-17. doi: 10.3233/JHD-170235. J Huntingtons Dis. 2017. PMID: 28339401 Free PMC article. Review.
-
Huntingtin polyQ Mutation Impairs the 17β-Estradiol/Neuroglobin Pathway Devoted to Neuron Survival.Mol Neurobiol. 2017 Oct;54(8):6634-6646. doi: 10.1007/s12035-016-0337-x. Epub 2016 Dec 12. Mol Neurobiol. 2017. PMID: 27957684
-
Dominant effects of the Huntington's disease HTT CAG repeat length are captured in gene-expression data sets by a continuous analysis mathematical modeling strategy.Hum Mol Genet. 2013 Aug 15;22(16):3227-38. doi: 10.1093/hmg/ddt176. Epub 2013 Apr 16. Hum Mol Genet. 2013. PMID: 23595883 Free PMC article.
-
Huntingtin gene evolution in Chordata and its peculiar features in the ascidian Ciona genus.BMC Genomics. 2006 Nov 8;7:288. doi: 10.1186/1471-2164-7-288. BMC Genomics. 2006. PMID: 17092333 Free PMC article.
References
-
- Martin JB, Gusella JF. Huntington's disease. Pathogenesis and management. N Engl J Med. 1986;315:1267–1276. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

