Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors
- PMID: 12297621
- PMCID: PMC130569
- DOI: 10.1073/pnas.162471999
Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors
Abstract
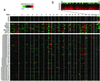
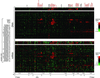
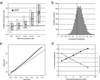
Genomic DNA copy number alterations are key genetic events in the development and progression of human cancers. Here we report a genome-wide microarray comparative genomic hybridization (array CGH) analysis of DNA copy number variation in a series of primary human breast tumors. We have profiled DNA copy number alteration across 6,691 mapped human genes, in 44 predominantly advanced, primary breast tumors and 10 breast cancer cell lines. While the overall patterns of DNA amplification and deletion corroborate previous cytogenetic studies, the high-resolution (gene-by-gene) mapping of amplicon boundaries and the quantitative analysis of amplicon shape provide significant improvement in the localization of candidate oncogenes. Parallel microarray measurements of mRNA levels reveal the remarkable degree to which variation in gene copy number contributes to variation in gene expression in tumor cells. Specifically, we find that 62% of highly amplified genes show moderately or highly elevated expression, that DNA copy number influences gene expression across a wide range of DNA copy number alterations (deletion, low-, mid- and high-level amplification), that on average, a 2-fold change in DNA copy number is associated with a corresponding 1.5-fold change in mRNA levels, and that overall, at least 12% of all the variation in gene expression among the breast tumors is directly attributable to underlying variation in gene copy number. These findings provide evidence that widespread DNA copy number alteration can lead directly to global deregulation of gene expression, which may contribute to the development or progression of cancer.
Figures

Similar articles
-
Impact of DNA amplification on gene expression patterns in breast cancer.Cancer Res. 2002 Nov 1;62(21):6240-5. Cancer Res. 2002. PMID: 12414653
-
High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays.Nat Genet. 1998 Oct;20(2):207-11. doi: 10.1038/2524. Nat Genet. 1998. PMID: 9771718
-
New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays.Cancer Res. 2001 Nov 15;61(22):8235-40. Cancer Res. 2001. PMID: 11719455
-
The 17q23 amplicon and breast cancer.Breast Cancer Res Treat. 2003 Apr;78(3):313-22. doi: 10.1023/a:1023081624133. Breast Cancer Res Treat. 2003. PMID: 12755490 Review.
-
Profiling breast cancer by array CGH.Breast Cancer Res Treat. 2003 Apr;78(3):289-98. doi: 10.1023/a:1023025506386. Breast Cancer Res Treat. 2003. PMID: 12755488 Review.
Cited by
-
Overexpression of TPD52L2 in HNSCC: prognostic significance and correlation with immune infiltrates.BMC Oral Health. 2024 Oct 7;24(1):1191. doi: 10.1186/s12903-024-04977-1. BMC Oral Health. 2024. PMID: 39375696 Free PMC article.
-
Performance enhancement of classifiers through Bio inspired feature selection methods for early detection of lung cancer from microarray genes.Heliyon. 2024 Aug 17;10(16):e36419. doi: 10.1016/j.heliyon.2024.e36419. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39262982 Free PMC article.
-
Allele-specific transcriptional effects of subclonal copy number alterations enable genotype-phenotype mapping in cancer cells.Nat Commun. 2024 Mar 20;15(1):2482. doi: 10.1038/s41467-024-46710-0. Nat Commun. 2024. PMID: 38509111 Free PMC article.
-
SIRT1 mediates breast cancer development and tumorigenesis controlled by estrogen-related receptor β.Breast Cancer. 2024 May;31(3):440-455. doi: 10.1007/s12282-024-01555-9. Epub 2024 Feb 29. Breast Cancer. 2024. PMID: 38421553
-
Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits.Front Vet Sci. 2024 Jan 11;10:1334434. doi: 10.3389/fvets.2023.1334434. eCollection 2023. Front Vet Sci. 2024. PMID: 38274664 Free PMC article. Review.
References
-
- Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Science. 1992;258:818–821. - PubMed
-
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O P. Genes Chromosomes Cancer. 1998;21:177–184. - PubMed
-
- Forozan F, Mahlamaki E H, Monni O, Chen Y, Veldman R, Jiang Y, Gooden G C, Ethier S P, Kallioniemi A, Kallioniemi O P. Cancer Res. 2000;60:4519–4525. - PubMed
-
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Genes Chromosomes Cancer. 1997;20:399–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

