An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP
- PMID: 12271126
- PMCID: PMC129685
- DOI: 10.1073/pnas.202200399
An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP
Abstract
In herpes simplex virus, lytic replication is initiated by the viral transactivator VP16 acting with cellular cofactors Oct-1 and HCF-1. Although this activator complex has been studied in detail, the role of HCF-1 remains elusive. Here, we show that HCF-1 contains an activation domain (HCF-1(AD)) required for maximal transactivation by VP16 and its cellular counterpart LZIP. Expression of the VP16 cofactor p300 augments HCF-1(AD) activity, suggesting a mechanism of synergy. Infection of cells lacking the HCF-1(AD) leads to reduced viral immediate-early gene expression and lowered viral titers. These findings underscore the importance of HCF-1 to herpes simplex virus replication and VP16 transactivation.
Figures
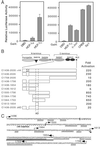
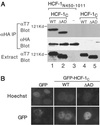
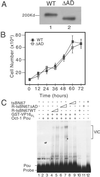

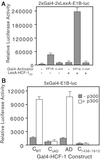
Similar articles
-
N-terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif.Proc Natl Acad Sci U S A. 2000 Sep 26;97(20):10757-62. doi: 10.1073/pnas.190062797. Proc Natl Acad Sci U S A. 2000. PMID: 10984507 Free PMC article.
-
Mutations in host cell factor 1 separate its role in cell proliferation from recruitment of VP16 and LZIP.Mol Cell Biol. 2000 Feb;20(3):919-28. doi: 10.1128/MCB.20.3.919-928.2000. Mol Cell Biol. 2000. PMID: 10629049 Free PMC article.
-
Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16.Nucleic Acids Res. 2000 Jun 15;28(12):2446-54. doi: 10.1093/nar/28.12.2446. Nucleic Acids Res. 2000. PMID: 10871379 Free PMC article.
-
The herpes simplex virus VP16-induced complex: the makings of a regulatory switch.Trends Biochem Sci. 2003 Jun;28(6):294-304. doi: 10.1016/S0968-0004(03)00088-4. Trends Biochem Sci. 2003. PMID: 12826401 Review.
-
The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation.Cold Spring Harb Symp Quant Biol. 1998;63:599-607. doi: 10.1101/sqb.1998.63.599. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384325 Review. No abstract available.
Cited by
-
The neuronal host cell factor-binding protein Zhangfei inhibits herpes simplex virus replication.J Virol. 2005 Dec;79(23):14708-18. doi: 10.1128/JVI.79.23.14708-14718.2005. J Virol. 2005. PMID: 16282471 Free PMC article.
-
HDAC3 selectively represses CREB3-mediated transcription and migration of metastatic breast cancer cells.Cell Mol Life Sci. 2010 Oct;67(20):3499-510. doi: 10.1007/s00018-010-0388-5. Epub 2010 May 15. Cell Mol Life Sci. 2010. PMID: 20473547 Free PMC article.
-
A novel protein, Luman/CREB3 recruitment factor, inhibits Luman activation of the unfolded protein response.Mol Cell Biol. 2008 Jun;28(12):3952-66. doi: 10.1128/MCB.01439-07. Epub 2008 Apr 7. Mol Cell Biol. 2008. PMID: 18391022 Free PMC article.
-
HCF-1 functions as a coactivator for the zinc finger protein Krox20.J Biol Chem. 2003 Dec 19;278(51):51116-24. doi: 10.1074/jbc.M303470200. Epub 2003 Oct 6. J Biol Chem. 2003. PMID: 14532282 Free PMC article.
-
Molecular cloning of Drosophila HCF reveals proteolytic processing and self-association of the encoded protein.J Cell Physiol. 2003 Feb;194(2):117-26. doi: 10.1002/jcp.10193. J Cell Physiol. 2003. PMID: 12494450 Free PMC article.
References
-
- Flint J, Shenk T. Annu Rev Genet. 1997;31:177–212. - PubMed
-
- Weir J P. Gene. 2001;271:117–130. - PubMed
-
- O'Hare P. Semin Virol. 1993;4:145–155.
-
- Herr W. Cold Spring Harbor Symp Quant Biol. 1998;63:599–607. - PubMed
-
- Wilson A C, Cleary M A, Lai J S, LaMarco K, Peterson M G, Herr W. Cold Spring Harbor Symp Quant Biol. 1993;58:167–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

