Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis
- PMID: 12208937
- PMCID: PMC136503
- DOI: 10.1128/jvi.76.19.9575-9587.2002
Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis
Abstract
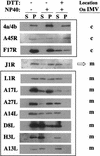
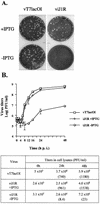
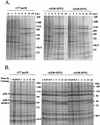
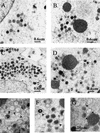
Vaccinia virus, a member of the poxvirus family, contains a conserved J1R open reading frame that encodes a late protein of 17.8 kDa. The 18-kDa J1R protein is associated mainly with the membrane fraction of intracellular mature virus particles. This study examines the biological function of J1R protein in the vaccinia virus life cycle. A recombinant vaccinia virus was constructed to conditionally express J1R protein in an isopropyl-beta-D-galactopyranoside (IPTG)-inducible manner. When J1R is not expressed during vaccinia virus infection, the virus titer is reduced approximately 100-fold. In contrast, J1R protein is not required for viral gene expression, as indicated by protein pulse-labeling. J1R protein is also not required for DNA processing, as the resolution of the concatemer junctions of replicated viral DNA was detected without IPTG. A deficiency of J1R protein caused a severe delay in the processing of p4a and p4b into mature core proteins 4a and 4b, indicating that J1R protein participates in virion morphogenesis. Infected cells grown in the absence of IPTG contained very few intracellular mature virions in the cytoplasm, and enlarged viroplasm structures accumulated with viral crescents attached at the periphery. Abundant intermediate membrane structures of abnormal shapes were observed, and many immature virions were either empty or partially filled, indicating that J1R protein is important for DNA packaging into immature virions. J1R protein also coimmunoprecipited with A45R protein in infected cells. In summary, these results indicate that vaccinia virus J1R is a membrane protein that is required for virus growth and plaque formation. J1R protein interacts with A45R protein and performs an important role during immature virion formation in cultured cells.
Figures





Similar articles
-
Vaccinia virus WR gene A5L is required for morphogenesis of mature virions.J Virol. 1999 Jun;73(6):4590-9. doi: 10.1128/JVI.73.6.4590-4599.1999. J Virol. 1999. PMID: 10233918 Free PMC article.
-
The vaccinia virus I1 protein is essential for the assembly of mature virions.J Virol. 1997 Dec;71(12):9285-94. doi: 10.1128/JVI.71.12.9285-9294.1997. J Virol. 1997. PMID: 9371587 Free PMC article.
-
Effects of a temperature sensitivity mutation in the J1R protein component of a complex required for vaccinia virus assembly.J Virol. 2005 Jul;79(13):8046-56. doi: 10.1128/JVI.79.13.8046-8056.2005. J Virol. 2005. PMID: 15956550 Free PMC article.
-
The exit of vaccinia virus from infected cells.Virus Res. 2004 Dec;106(2):189-97. doi: 10.1016/j.virusres.2004.08.015. Virus Res. 2004. PMID: 15567497 Review.
-
In a nutshell: structure and assembly of the vaccinia virion.Adv Virus Res. 2006;66:31-124. doi: 10.1016/S0065-3527(06)66002-8. Adv Virus Res. 2006. PMID: 16877059 Review.
Cited by
-
From crescent to mature virion: vaccinia virus assembly and maturation.Viruses. 2014 Oct 7;6(10):3787-808. doi: 10.3390/v6103787. Viruses. 2014. PMID: 25296112 Free PMC article. Review.
-
Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus.J Virol. 2006 Nov;80(21):10615-23. doi: 10.1128/JVI.01452-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928742 Free PMC article.
-
Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles.J Virol. 2006 Mar;80(5):2127-40. doi: 10.1128/JVI.80.5.2127-2140.2006. J Virol. 2006. PMID: 16474121 Free PMC article.
-
Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins.J Virol. 2005 Jun;79(11):7146-61. doi: 10.1128/JVI.79.11.7146-7161.2005. J Virol. 2005. PMID: 15890954 Free PMC article.
-
Vaccinia virus morphogenesis: a13 phosphoprotein is required for assembly of mature virions.J Virol. 2004 Aug;78(16):8885-901. doi: 10.1128/JVI.78.16.8885-8901.2004. J Virol. 2004. PMID: 15280497 Free PMC article.
References
-
- Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

