Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53-independent cell death
- PMID: 12205095
- PMCID: PMC1201509
- DOI: 10.1074/jbc.M207662200
Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53-independent cell death
Abstract
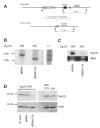
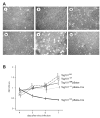
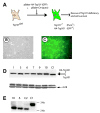
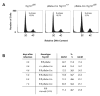
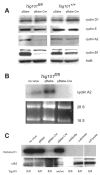
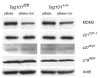
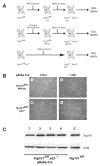
The tumor susceptibility gene 101 (Tsg101) was originally discovered in a screen for potential tumor suppressors using insertional mutagenesis in immortalized fibroblasts. To investigate essential functions of this gene in cell growth and neoplastic transformation, we derived primary mouse embryonic fibroblasts from Tsg101 conditional knockout mice. Expression of Cre recombinase from a retroviral vector efficiently down-regulated Tsg101. The deletion of Tsg101 caused growth arrest and cell death but did not result in increased proliferation and cellular transformation. Inactivation of p53 had no influence on the deleterious phenotype, but Tsg101(-/-) cells were rescued through expression of exogenous Tsg101. Fluorescence-activated cell sorting, proliferation assays, and Western blot analysis of crucial regulators of the cell cycle revealed that Tsg101 deficiency resulted in growth arrest at the G(1)/S transition through inactivation of cyclin-dependent kinase 2. As a consequence, DNA replication was not initiated in Tsg101-deficient cells. Our results clearly demonstrate that Tsg101 is not a primary tumor suppressor in mouse embryonic fibroblasts. However, the protein is crucial for cell proliferation and cell survival.
Figures

Similar articles
-
A knockout of the Tsg101 gene leads to decreased expression of ErbB receptor tyrosine kinases and induction of autophagy prior to cell death.PLoS One. 2012;7(3):e34308. doi: 10.1371/journal.pone.0034308. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479596 Free PMC article.
-
Cell cycle arrest and cell death are controlled by p53-dependent and p53-independent mechanisms in Tsg101-deficient cells.J Biol Chem. 2004 Aug 20;279(34):35984-94. doi: 10.1074/jbc.M400408200. Epub 2004 Jun 21. J Biol Chem. 2004. PMID: 15210712 Free PMC article.
-
p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101.Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):1859-64. doi: 10.1073/pnas.98.4.1859. Proc Natl Acad Sci U S A. 2001. PMID: 11172041 Free PMC article.
-
Role of TSG101 in cancer.Front Biosci (Landmark Ed). 2013 Jan 1;18(1):279-88. doi: 10.2741/4099. Front Biosci (Landmark Ed). 2013. PMID: 23276921 Review.
-
[Molecular mechanisms controlling the cell cycle: fundamental aspects and implications for oncology].Cancer Radiother. 2001 Apr;5(2):109-29. doi: 10.1016/s1278-3218(01)00087-7. Cancer Radiother. 2001. PMID: 11355576 Review. French.
Cited by
-
Tsg101 is essential for cell growth, proliferation, and cell survival of embryonic and adult tissues.Mol Cell Biol. 2003 Jan;23(1):150-62. doi: 10.1128/MCB.23.1.150-162.2003. Mol Cell Biol. 2003. PMID: 12482969 Free PMC article.
-
TSG101, identified by screening a cancer cDNA library and soft agar assay, promotes cell proliferation in human lung cancer.Mol Biol Rep. 2010 Jul;37(6):2829-38. doi: 10.1007/s11033-009-9835-5. Epub 2009 Sep 29. Mol Biol Rep. 2010. PMID: 19787439
-
The role of the endosomal sorting complexes required for transport (ESCRT) in tumorigenesis.Mol Membr Biol. 2014 Jun;31(4):111-9. doi: 10.3109/09687688.2014.894210. Epub 2014 Mar 18. Mol Membr Biol. 2014. PMID: 24641493 Free PMC article. Review.
-
A mammary-specific, long-range deletion on mouse chromosome 11 accelerates Brca1-associated mammary tumorigenesis.Neoplasia. 2008 Dec;10(12):1325-34. doi: 10.1593/neo.08524. Neoplasia. 2008. PMID: 19048111 Free PMC article.
-
A knockout of the Tsg101 gene leads to decreased expression of ErbB receptor tyrosine kinases and induction of autophagy prior to cell death.PLoS One. 2012;7(3):e34308. doi: 10.1371/journal.pone.0034308. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479596 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

