Dnmt3L is a transcriptional repressor that recruits histone deacetylase
- PMID: 12202768
- PMCID: PMC137431
- DOI: 10.1093/nar/gkf509
Dnmt3L is a transcriptional repressor that recruits histone deacetylase
Abstract
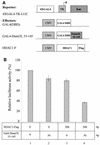
The Dnmt3L protein belongs to the Dnmt3 family of DNA methyltransferases by virtue of its sequence homology in the plant homeodomain (PHD)-like motif. Dnmt3L is essential for the establishment of maternal genomic imprints and, given its lack of key methyltransferase motifs, is more likely to act as a regulator of methylation rather than as an enzyme that methylates DNA. Here, we show that Dnmt3L, like Dnmt3a and Dnmt3b, interacts both in vitro and in vivo with the histone deacetylase HDAC1. Consistent with the binding to a deacetylase, Dnmt3L purifies histone deacetylase activity from nuclear extracts. We find that Dnmt3L can repress transcription and that this repression is dependent on HDAC1 and is relieved by treatment with the HDAC inhibitor trichostatin A. Binding of Dnmt3L to HDAC1 as well as its repressive function require the PHD-like motif. Our results indicate that Dnmt3L plays a role in transcriptional regulation and that recruitment of the HDAC repressive machinery is a shared and conserved feature of the Dnmt3 family. The fact that, despite the absence of a methyltransferase domain, Dnmt3L retains the capacity to contact deacetylase further substantiates the notion that the Dnmts can repress transcription independently of their methylating activities.
Figures
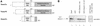
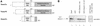




Similar articles
-
Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity.Nucleic Acids Res. 2002 Aug 15;30(16):3602-8. doi: 10.1093/nar/gkf474. Nucleic Acids Res. 2002. PMID: 12177302 Free PMC article.
-
Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription.J Virol. 1999 Jul;73(7):5688-97. doi: 10.1128/JVI.73.7.5688-5697.1999. J Virol. 1999. PMID: 10364319 Free PMC article.
-
Ectopic DNMT3L triggers assembly of a repressive complex for retroviral silencing in somatic cells.J Virol. 2014 Sep;88(18):10680-95. doi: 10.1128/JVI.01176-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24991018 Free PMC article.
-
Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription.EMBO J. 2001 May 15;20(10):2536-44. doi: 10.1093/emboj/20.10.2536. EMBO J. 2001. PMID: 11350943 Free PMC article.
-
The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome.Genes (Basel). 2019 Feb 23;10(2):172. doi: 10.3390/genes10020172. Genes (Basel). 2019. PMID: 30813436 Free PMC article. Review.
Cited by
-
Epigenetic modifications affect Dnmt3L expression.Biochem J. 2004 Jun 15;380(Pt 3):705-13. doi: 10.1042/BJ20040067. Biochem J. 2004. PMID: 15015937 Free PMC article.
-
Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs.Curr Med Chem. 2010;17(33):4052-71. doi: 10.2174/092986710793205372. Curr Med Chem. 2010. PMID: 20939822 Free PMC article. Review.
-
Immunomodulatory Properties of Human Breast Milk: MicroRNA Contents and Potential Epigenetic Effects.Biomedicines. 2022 May 24;10(6):1219. doi: 10.3390/biomedicines10061219. Biomedicines. 2022. PMID: 35740242 Free PMC article. Review.
-
Metabolism and the Epigenome: A Dynamic Relationship.Trends Biochem Sci. 2020 Sep;45(9):731-747. doi: 10.1016/j.tibs.2020.04.002. Epub 2020 May 6. Trends Biochem Sci. 2020. PMID: 32387193 Free PMC article. Review.
-
The Role of Epigenetics in Autoimmune/Inflammatory Disease.Front Immunol. 2019 Jul 4;10:1525. doi: 10.3389/fimmu.2019.01525. eCollection 2019. Front Immunol. 2019. PMID: 31333659 Free PMC article. Review.
References
-
- Jones P.A. and Takai,D. (2001) Cancer epigenetics comes of age. Science, 293, 1068–1070. - PubMed
-
- Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. - PubMed
-
- Jaenisch R. (1997) DNA methylation and imprinting: why bother? Trends Genet., 13, 323–329. - PubMed
-
- Surani M.A. (2001) Reprogramming of genome function through epigenetic inheritance. Nature, 414, 122–128. - PubMed
-
- Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

