The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway
- PMID: 12154127
- PMCID: PMC186409
- DOI: 10.1101/gad.996502
The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway
Abstract
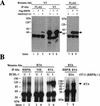
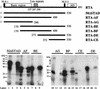
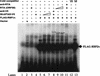
The RTA protein of the Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is responsible for the switch from latency to lytic replication, a reaction essential for viral spread and KS pathogenesis. RTA is a sequence-specific transcriptional activator, but the diversity of its target sites suggests it may act via interaction with host DNA-binding proteins as well. Here we show that KSHV RTA interacts with the RBP-Jkappa protein, the primary target of the Notch signaling pathway. This interaction targets RTA to RBP-Jkappa recognition sites on DNA and results in the replacement of RBP-Jkappa's intrinsic repressive action with activation mediated by the C-terminal domain of RTA. Mutation of such sites in target promoters strongly impairs RTA responsiveness. Similarly, such target genes are induced poorly or not at all by RTA in fibroblasts derived from RBP-Jkappa(-/-) mice, a defect that can be reversed by expression of RBP-Jkappa. In vitro, RTA binds to two adjacent regions of RBP-Jkappa, one of which is identical to the central repression domain that binds the Notch effector fragment. These results indicate that KSHV has evolved a ligand-independent mechanism for constitutive activation of the Notch pathway as a part of its strategy for reactivation from latency.
Figures
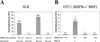




Similar articles
-
Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling.Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8490-5. doi: 10.1073/pnas.1432843100. Epub 2003 Jun 27. Proc Natl Acad Sci U S A. 2003. PMID: 12832621 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway.J Virol. 2005 Mar;79(6):3468-78. doi: 10.1128/JVI.79.6.3468-3478.2005. J Virol. 2005. PMID: 15731241 Free PMC article.
-
The interaction between KSHV RTA and cellular RBP-Jkappa and their subsequent DNA binding are not sufficient for activation of RBP-Jkappa.Virus Res. 2008 Jan;131(1):1-7. doi: 10.1016/j.virusres.2007.07.019. Epub 2007 Sep 11. Virus Res. 2008. PMID: 17850910 Free PMC article.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
-
The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression.Oncogene. 2003 Aug 11;22(33):5150-63. doi: 10.1038/sj.onc.1206555. Oncogene. 2003. PMID: 12910252 Review.
Cited by
-
Number of and distance between response elements in Kaposi's sarcoma-associated herpesvirus ORF57 promoter influence its activation by replication and transcription activator and its repression by interferon regulatory factor 7.Arch Virol. 2010 Mar;155(3):361-6. doi: 10.1007/s00705-009-0576-5. Epub 2009 Dec 29. Arch Virol. 2010. PMID: 20039088 Free PMC article.
-
An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer.PLoS Biol. 2007 Jan;5(1):e9. doi: 10.1371/journal.pbio.0050009. PLoS Biol. 2007. PMID: 17194214 Free PMC article.
-
Modulation of human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus replication and transcription activator transactivation by interferon regulatory factor 7.J Virol. 2005 Feb;79(4):2420-31. doi: 10.1128/JVI.79.4.2420-2431.2005. J Virol. 2005. PMID: 15681443 Free PMC article.
-
Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling.Mol Syst Biol. 2013 Oct 1;9:692. doi: 10.1038/msb.2013.50. Mol Syst Biol. 2013. PMID: 24084807 Free PMC article.
-
KSHV reactivation from latency requires Pim-1 and Pim-3 kinases to inactivate the latency-associated nuclear antigen LANA.PLoS Pathog. 2009 Mar;5(3):e1000324. doi: 10.1371/journal.ppat.1000324. Epub 2009 Mar 6. PLoS Pathog. 2009. PMID: 19266083 Free PMC article.
References
-
- Ambroziak JA, Blackbourn DJ, Herndier BG, Glogau RG, Gullett JH, McDonald AR, Lennette ET, Levy JA. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science. 1995;268:582–583. - PubMed
-
- Aster J, Pear W. Notch signaling in leukemia. Curr Opin Hematol. 2001;8:237–244. - PubMed
-
- Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, Asch AS, Cesarman E, Gershengorn MC, Mesri EA, et al. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. - PubMed
-
- Blackbourn DJ, Lennette E, Klencke B, Moses A, Chandran B, Weinstein M, Glogau RG, Witte MH, Way DL, Kutzkey T, et al. The restricted cellular host range of human herpesvirus 8. AIDS. 2000;14:1123–1133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
