Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control
- PMID: 12154125
- PMCID: PMC186427
- DOI: 10.1101/gad.1002002
Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control
Abstract
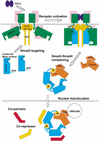
Smad3 transduces the signals of TGF-betas, coupling transmembrane receptor kinase activation to transcriptional control. The membrane-associated molecule SARA (Smad Anchor for Receptor Activation) recruits Smad3 for phosphorylation by the receptor kinase. Upon phosphorylation, Smad3 dissociates from SARA and enters the nucleus, in which its transcriptional activity can be repressed by Ski. Here, we show that SARA and Ski recognize specifically the monomeric and trimeric forms of Smad3, respectively. Thus, trimerization of Smad3, induced by phosphorylation, simultaneously activates the TGF-beta signal by driving Smad3 dissociation from SARA and sets up the negative feedback mechanism by Ski. Structural models of the Smad3/SARA/receptor kinase complex and Smad3/Ski complex provide insights into the molecular basis of regulation.
Figures


Comment in
-
From mono- to oligo-Smads: the heart of the matter in TGF-beta signal transduction.Genes Dev. 2002 Aug 1;16(15):1867-71. doi: 10.1101/gad.1016802. Genes Dev. 2002. PMID: 12154118 No abstract available.
Similar articles
-
From mono- to oligo-Smads: the heart of the matter in TGF-beta signal transduction.Genes Dev. 2002 Aug 1;16(15):1867-71. doi: 10.1101/gad.1016802. Genes Dev. 2002. PMID: 12154118 No abstract available.
-
Two short segments of Smad3 are important for specific interaction of Smad3 with c-Ski and SnoN.J Biol Chem. 2003 Jan 3;278(1):531-6. doi: 10.1074/jbc.C200596200. Epub 2002 Nov 7. J Biol Chem. 2003. PMID: 12426322
-
Structural basis of Smad2 recognition by the Smad anchor for receptor activation.Science. 2000 Jan 7;287(5450):92-7. doi: 10.1126/science.287.5450.92. Science. 2000. PMID: 10615055
-
TGF-beta signaling by Smad proteins.Adv Immunol. 2000;75:115-57. doi: 10.1016/s0065-2776(00)75003-6. Adv Immunol. 2000. PMID: 10879283 Review.
-
[Role of Ski/SnoN protein in the regulation of TGF-beta signal pathway].Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003 Apr;25(2):233-6. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003. PMID: 12905729 Review. Chinese.
Cited by
-
Transforming growth factor-β in tumour development.Front Mol Biosci. 2022 Oct 4;9:991612. doi: 10.3389/fmolb.2022.991612. eCollection 2022. Front Mol Biosci. 2022. PMID: 36267157 Free PMC article. Review.
-
Structural Basis of Intracellular TGF-β Signaling: Receptors and Smads.Cold Spring Harb Perspect Biol. 2016 Nov 1;8(11):a022111. doi: 10.1101/cshperspect.a022111. Cold Spring Harb Perspect Biol. 2016. PMID: 27549117 Free PMC article. Review.
-
Interaction with Smad4 is indispensable for suppression of BMP signaling by c-Ski.Mol Biol Cell. 2004 Mar;15(3):963-72. doi: 10.1091/mbc.e03-07-0478. Epub 2003 Dec 29. Mol Biol Cell. 2004. PMID: 14699069 Free PMC article.
-
Functional Testing of Bone Morphogenetic Protein (BMP) Pathway Variants Identified on Whole-Exome Sequencing in a Patient with Delayed-Onset Fibrodysplasia Ossificans Progressiva (FOP) Using ACVR1R206H -Specific Human Cellular and Zebrafish Models.J Bone Miner Res. 2022 Nov;37(11):2058-2076. doi: 10.1002/jbmr.4711. Epub 2022 Nov 15. J Bone Miner Res. 2022. PMID: 36153796 Free PMC article.
-
Expressed protein ligation: a resourceful tool to study protein structure and function.Cell Mol Life Sci. 2009 Dec;66(24):3909-22. doi: 10.1007/s00018-009-0122-3. Cell Mol Life Sci. 2009. PMID: 19685006 Free PMC article. Review.
References
-
- Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. - PubMed
-
- Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with Smads. J Biol Chem. 1999;274:35269–35277. - PubMed
-
- Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–243. - PubMed
-
- Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor β in human disease. New Eng J Med. 2000;342:1350–1358. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
