Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex
- PMID: 12101232
- PMCID: PMC133947
- DOI: 10.1128/MCB.22.15.5367-5379.2002
Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex
Abstract
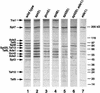
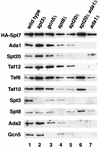
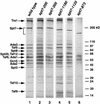
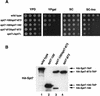
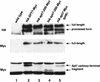
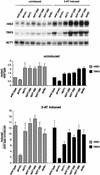
The Saccharomyces cerevisiae SAGA complex is required for the normal transcription of a large number of genes. Complex integrity depends on three core subunits, Spt7, Spt20, and Ada1. We have investigated the role of Spt7 in the assembly and function of SAGA. Our results show that Spt7 is important in controlling the levels of the other core subunits and therefore of SAGA. In addition, partial SAGA complexes containing Spt7 can be assembled in the absence of both Spt20 and Ada1. Through biochemical and genetic analyses of a series of spt7 deletion mutants, we have identified a region of Spt7 required for interaction with the SAGA component Spt8. An adjacent Spt7 domain was found to be required for a processed form of Spt7 that is present in a previously identified altered form of SAGA called SLIK, SAGA(alt), or SALSA. Analysis of an spt7 mutant with greatly reduced levels of SLIK/SAGA(alt)/SALSA suggests a subtle role for this complex in transcription that may be redundant with a subset of SAGA functions.
Figures



Similar articles
-
Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters.Mol Cell Biol. 2000 Jan;20(2):634-47. doi: 10.1128/MCB.20.2.634-647.2000. Mol Cell Biol. 2000. PMID: 10611242 Free PMC article.
-
Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction.Mol Cell Biol. 1999 Jan;19(1):86-98. doi: 10.1128/MCB.19.1.86. Mol Cell Biol. 1999. PMID: 9858534 Free PMC article.
-
SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription.Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11622-7. doi: 10.1073/pnas.182021199. Epub 2002 Aug 19. Proc Natl Acad Sci U S A. 2002. PMID: 12186975 Free PMC article.
-
C-terminal processing of yeast Spt7 occurs in the absence of functional SAGA complex.BMC Biochem. 2007 Aug 8;8:16. doi: 10.1186/1471-2091-8-16. BMC Biochem. 2007. PMID: 17686179 Free PMC article.
-
SAGA-CORE subunit Spt7 is required for correct Ubp8 localization, chromatin association and deubiquitinase activity.Epigenetics Chromatin. 2020 Oct 28;13(1):46. doi: 10.1186/s13072-020-00367-3. Epigenetics Chromatin. 2020. PMID: 33115507 Free PMC article.
Cited by
-
STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation.Mol Cell Biol. 2008 Jan;28(1):108-21. doi: 10.1128/MCB.01402-07. Epub 2007 Oct 29. Mol Cell Biol. 2008. PMID: 17967894 Free PMC article.
-
Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3.Genes Dev. 2008 Nov 1;22(21):2994-3006. doi: 10.1101/gad.1724408. Genes Dev. 2008. PMID: 18981477 Free PMC article.
-
The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species.Microbiol Mol Biol Rev. 2016 Feb 10;80(1):205-327. doi: 10.1128/MMBR.00040-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2016. PMID: 26864432 Free PMC article. Review.
-
The SAGA complex, together with transcription factors and the endocytic protein Rvs167p, coordinates the reprofiling of gene expression in response to changes in sterol composition in Saccharomyces cerevisiae.Mol Biol Cell. 2017 Oct 1;28(20):2637-2649. doi: 10.1091/mbc.E17-03-0169. Epub 2017 Aug 2. Mol Biol Cell. 2017. PMID: 28768829 Free PMC article.
-
Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2013 Apr 30;110(18):7318-23. doi: 10.1073/pnas.1302490110. Epub 2013 Apr 15. Proc Natl Acad Sci U S A. 2013. PMID: 23589851 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. Greene Publishing Associates/Wiley-Interscience, New York, N.Y.
-
- Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2001. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
