MicroRNAs in plants
- PMID: 12101121
- PMCID: PMC186362
- DOI: 10.1101/gad.1004402
MicroRNAs in plants
Erratum in
- Genes Dev 2002 Sep 1;16(17):2313
Abstract
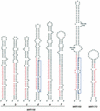
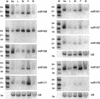
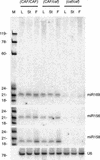
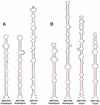
MicroRNAs (miRNAs) are an extensive class of ~22-nucleotide noncoding RNAs thought to regulate gene expression in metazoans. We find that miRNAs are also present in plants, indicating that this class of noncoding RNA arose early in eukaryotic evolution. In this paper 16 Arabidopsis miRNAs are described, many of which have differential expression patterns in development. Eight are absolutely conserved in the rice genome. The plant miRNA loci potentially encode stem-loop precursors similar to those processed by Dicer (a ribonuclease III) in animals. Mutation of an Arabidopsis Dicer homolog, CARPEL FACTORY, prevents the accumulation of miRNAs, showing that similar mechanisms direct miRNA processing in plants and animals. The previously described roles of CARPEL FACTORY in the development of Arabidopsis embryos, leaves, and floral meristems suggest that the miRNAs could play regulatory roles in the development of plants as well as animals.
Figures
Similar articles
-
CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana.Curr Biol. 2002 Sep 3;12(17):1484-95. doi: 10.1016/s0960-9822(02)01017-5. Curr Biol. 2002. PMID: 12225663 Free PMC article.
-
Computational identification of plant microRNAs and their targets, including a stress-induced miRNA.Mol Cell. 2004 Jun 18;14(6):787-99. doi: 10.1016/j.molcel.2004.05.027. Mol Cell. 2004. PMID: 15200956
-
Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes.Proc Natl Acad Sci U S A. 2004 Aug 3;101(31):11511-6. doi: 10.1073/pnas.0404025101. Epub 2004 Jul 22. Proc Natl Acad Sci U S A. 2004. PMID: 15272084 Free PMC article.
-
Expression of microRNAs and its regulation in plants.Semin Cell Dev Biol. 2010 Oct;21(8):790-7. doi: 10.1016/j.semcdb.2010.03.012. Epub 2010 Apr 18. Semin Cell Dev Biol. 2010. PMID: 20403450 Free PMC article. Review.
-
Conservation and divergence in plant microRNAs.Plant Mol Biol. 2012 Sep;80(1):3-16. doi: 10.1007/s11103-011-9829-2. Epub 2011 Oct 14. Plant Mol Biol. 2012. PMID: 21996939 Review.
Cited by
-
miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment.Planta. 2016 Jul;244(1):231-43. doi: 10.1007/s00425-016-2505-7. Epub 2016 Apr 4. Planta. 2016. PMID: 27040841 Free PMC article.
-
Grass microRNA gene paleohistory unveils new insights into gene dosage balance in subgenome partitioning after whole-genome duplication.Plant Cell. 2012 May;24(5):1776-92. doi: 10.1105/tpc.112.095752. Epub 2012 May 15. Plant Cell. 2012. PMID: 22589464 Free PMC article.
-
miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets.PLoS One. 2012;7(8):e42390. doi: 10.1371/journal.pone.0042390. Epub 2012 Aug 1. PLoS One. 2012. PMID: 22870325 Free PMC article.
-
New insight into the molecular mechanism of miR482/2118 during plant resistance to pathogens.Front Plant Sci. 2022 Oct 28;13:1026762. doi: 10.3389/fpls.2022.1026762. eCollection 2022. Front Plant Sci. 2022. PMID: 36388487 Free PMC article. Review.
-
Current Status and Potential of RNA Interference for the Management of Tomato Spotted Wilt Virus and Thrips Vectors.Pathogens. 2021 Mar 9;10(3):320. doi: 10.3390/pathogens10030320. Pathogens. 2021. PMID: 33803131 Free PMC article. Review.
References
-
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Aravin AA, Naumova NM, Tulin AA, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in Drosophila melanogastergermline. Curr Biol. 2001;11:1017–1027. - PubMed
-
- Bender J. A vicious cycle: RNA silencing and DNA methylation in plants. Cell. 2001;106:129–132. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:295–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
