Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells
- PMID: 12096193
- PMCID: PMC126604
- DOI: 10.1073/pnas.152327299
Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells
Abstract
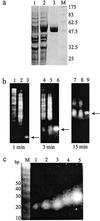
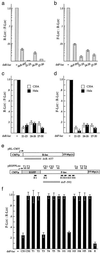
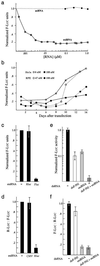
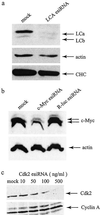
Small interfering RNA (siRNA) has become a powerful tool for selectively silencing gene expression in cultured mammalian cells. Because different siRNAs of the same gene have variable silencing capacities, RNA interference with synthetic siRNA is inefficient and cost intensive, especially for functional genomic studies. Here we report the use of Escherichia coli RNase III to cleave double-stranded RNA (dsRNA) into endoribonuclease-prepared siRNA (esiRNA) that can target multiple sites within an mRNA. esiRNA recapitulates the potent and specific inhibition by long dsRNA in Drosophila S2 cells. In contrast to long dsRNA, esiRNA mediates effective RNA interference without apparent nonspecific effect in cultured mammalian cells. We found that sequence-specific interference by esiRNA and the nonspecific IFN response activated by long dsRNA are independent pathways in mammalian cells. esiRNA works by eliciting the destruction of its cognate mRNA. Because of its simplicity and potency, this approach is useful for analysis of mammalian gene functions.
Figures

Similar articles
-
RNA interference (RNAi) with RNase III-prepared siRNAs.Methods Mol Biol. 2004;252:471-82. doi: 10.1385/1-59259-746-7:471. Methods Mol Biol. 2004. PMID: 15017072
-
Coexpression of Escherichia coli RNase III in silkworm cells improves the efficiency of RNA interference induced by long hairpin dsRNAs.Insect Sci. 2013 Feb;20(1):69-77. doi: 10.1111/j.1744-7917.2012.01569.x. Epub 2012 Nov 6. Insect Sci. 2013. PMID: 23955827
-
Endoribonuclease-prepared short interfering RNAs induce effective and specific inhibition of human immunodeficiency virus type 1 replication.J Virol. 2007 Oct;81(19):10680-6. doi: 10.1128/JVI.00950-07. Epub 2007 Jul 25. J Virol. 2007. PMID: 17652404 Free PMC article.
-
A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst.Cell. 2001 Nov 16;107(4):415-8. doi: 10.1016/s0092-8674(01)00581-5. Cell. 2001. PMID: 11719182 Review.
-
The multifaceted roles of the RNA processing enzyme ribonuclease III.Indian J Biochem Biophys. 1996 Aug;33(4):253-60. Indian J Biochem Biophys. 1996. PMID: 8936814 Review.
Cited by
-
Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding.PLoS Genet. 2012 Sep;8(9):e1002982. doi: 10.1371/journal.pgen.1002982. Epub 2012 Sep 27. PLoS Genet. 2012. PMID: 23028375 Free PMC article.
-
Progress in the use of RNA interference as a therapy for chronic hepatitis B virus infection.Genome Med. 2010 Apr 28;2(4):28. doi: 10.1186/gm149. Genome Med. 2010. PMID: 20429960 Free PMC article.
-
Kank attenuates actin remodeling by preventing interaction between IRSp53 and Rac1.J Cell Biol. 2009 Jan 26;184(2):253-67. doi: 10.1083/jcb.200805147. J Cell Biol. 2009. PMID: 19171758 Free PMC article.
-
Silencing viruses by RNA interference.Microbes Infect. 2005 Apr;7(4):767-75. doi: 10.1016/j.micinf.2005.02.003. Epub 2005 Mar 13. Microbes Infect. 2005. PMID: 15820151 Free PMC article. Review.
-
Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference.Mol Biol Cell. 2005 Jul;16(7):3187-99. doi: 10.1091/mbc.e05-02-0167. Epub 2005 Apr 20. Mol Biol Cell. 2005. PMID: 15843429 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

