A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure
- PMID: 12095421
- PMCID: PMC119854
- DOI: 10.1186/1471-2105-3-18
A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure
Abstract
Background: Covariance models (CMs) are probabilistic models of RNA secondary structure, analogous to profile hidden Markov models of linear sequence. The dynamic programming algorithm for aligning a CM to an RNA sequence of length N is O(N3) in memory. This is only practical for small RNAs.
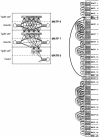
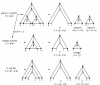
Results: I describe a divide and conquer variant of the alignment algorithm that is analogous to memory-efficient Myers/Miller dynamic programming algorithms for linear sequence alignment. The new algorithm has an O(N2 log N) memory complexity, at the expense of a small constant factor in time.
Conclusions: Optimal ribosomal RNA structural alignments that previously required up to 150 GB of memory now require less than 270 MB.
Figures

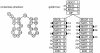
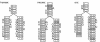
Similar articles
-
Reduced space hidden Markov model training.Bioinformatics. 1998 Jun;14(5):401-6. doi: 10.1093/bioinformatics/14.5.401. Bioinformatics. 1998. PMID: 9682053
-
Pair hidden Markov models on tree structures.Bioinformatics. 2003;19 Suppl 1:i232-40. doi: 10.1093/bioinformatics/btg1032. Bioinformatics. 2003. PMID: 12855464
-
Reduced space sequence alignment.Comput Appl Biosci. 1997 Feb;13(1):45-53. doi: 10.1093/bioinformatics/13.1.45. Comput Appl Biosci. 1997. PMID: 9088708
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
The art of editing RNA structural alignments.Methods Mol Biol. 2014;1097:379-94. doi: 10.1007/978-1-62703-709-9_17. Methods Mol Biol. 2014. PMID: 24639168 Review.
Cited by
-
Decreasing the number of false positives in sequence classification.BMC Genomics. 2010 Dec 22;11 Suppl 5(Suppl 5):S10. doi: 10.1186/1471-2164-11-S5-S10. BMC Genomics. 2010. PMID: 21210966 Free PMC article.
-
Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems.Appl Environ Microbiol. 2009 Jun;75(12):4035-45. doi: 10.1128/AEM.00515-09. Epub 2009 Apr 17. Appl Environ Microbiol. 2009. PMID: 19376903 Free PMC article.
-
NCBI Reference Sequences: current status, policy and new initiatives.Nucleic Acids Res. 2009 Jan;37(Database issue):D32-6. doi: 10.1093/nar/gkn721. Epub 2008 Oct 16. Nucleic Acids Res. 2009. PMID: 18927115 Free PMC article.
-
Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T.PLoS Genet. 2008 May 30;4(5):e1000087. doi: 10.1371/journal.pgen.1000087. PLoS Genet. 2008. PMID: 18516288 Free PMC article.
-
Computational identification of functional RNA homologs in metagenomic data.RNA Biol. 2013 Jul;10(7):1170-9. doi: 10.4161/rna.25038. Epub 2013 May 20. RNA Biol. 2013. PMID: 23722291 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

