Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription
- PMID: 12077340
- PMCID: PMC139777
- DOI: 10.1128/MCB.22.14.5114-5127.2002
Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription
Erratum in
- Mol Cell Biol 2002 Sep;22(17):6318
Abstract
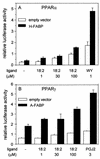
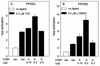
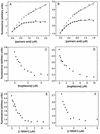
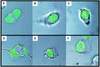
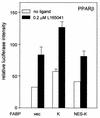
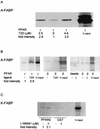
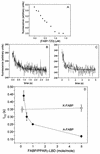
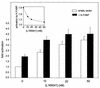
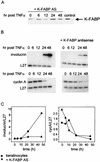
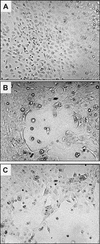
Lipophilic compounds such as retinoic acid and long-chain fatty acids regulate gene transcription by activating nuclear receptors such as retinoic acid receptors (RARs) and peroxisome proliferator-activated receptors (PPARs). These compounds also bind in cells to members of the family of intracellular lipid binding proteins, which includes cellular retinoic acid-binding proteins (CRABPs) and fatty acid binding proteins (FABPs). We previously reported that CRABP-II enhances the transcriptional activity of RAR by directly targeting retinoic acid to the receptor. Here, potential functional cooperation between FABPs and PPARs in regulating the transcriptional activities of their common ligands was investigated. We show that adipocyte FABP and keratinocyte FABP (A-FABP and K-FABP, respectively) selectively enhance the activities of PPARgamma and PPARbeta, respectively, and that these FABPs massively relocate to the nucleus in response to selective ligands for the PPAR isotype which they activate. We show further that A-FABP and K-FABP interact directly with PPARgamma and PPARbeta and that they do so in a receptor- and ligand-selective manner. Finally, the data demonstrate that the presence of high levels of K-FABP in keratinocytes is essential for PPARbeta-mediated induction of differentiation of these cells. Taken together, the data establish that A-FABP and K-FABP govern the transcriptional activities of their ligands by targeting them to cognate PPARs in the nucleus, thereby enabling PPARs to exert their biological functions.
Figures
Similar articles
-
A novel method for analysis of nuclear receptor function at natural promoters: peroxisome proliferator-activated receptor gamma agonist actions on aP2 gene expression detected using branched DNA messenger RNA quantitation.Mol Endocrinol. 1999 Mar;13(3):410-7. doi: 10.1210/mend.13.3.0246. Mol Endocrinol. 1999. PMID: 10076998
-
Expression of peroxisome proliferator-activated receptor gamma (PPARgamma) in human transitional bladder cancer and its role in inducing cell death.Neoplasia. 1999 Oct;1(4):330-9. doi: 10.1038/sj.neo.7900050. Neoplasia. 1999. PMID: 10935488 Free PMC article.
-
Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors alpha - and gamma-mediated gene expression via liver fatty acid binding protein: a signaling path to the nucleus.Proc Natl Acad Sci U S A. 2001 Feb 27;98(5):2323-8. doi: 10.1073/pnas.051619898. Epub 2001 Feb 20. Proc Natl Acad Sci U S A. 2001. PMID: 11226238 Free PMC article.
-
Cellular binding proteins for fatty acids and retinoids: similar or specialized functions?Mol Cell Biochem. 1993 Jun 9-23;123(1-2):191-202. doi: 10.1007/BF01076492. Mol Cell Biochem. 1993. PMID: 8232263 Review.
-
Cytoplasmic fatty acid-binding proteins: emerging roles in metabolism and atherosclerosis.Curr Opin Lipidol. 2002 Apr;13(2):141-7. doi: 10.1097/00041433-200204000-00005. Curr Opin Lipidol. 2002. PMID: 11891416 Review.
Cited by
-
Zinc finger protein 407 overexpression upregulates PPAR target gene expression and improves glucose homeostasis in mice.Am J Physiol Endocrinol Metab. 2016 Nov 1;311(5):E869-E880. doi: 10.1152/ajpendo.00234.2016. Epub 2016 Sep 13. Am J Physiol Endocrinol Metab. 2016. PMID: 27624101 Free PMC article.
-
Role of the Peroxisome Proliferator Activated Receptors in Hypertension.Circ Res. 2021 Apr 2;128(7):1021-1039. doi: 10.1161/CIRCRESAHA.120.318062. Epub 2021 Apr 1. Circ Res. 2021. PMID: 33793338 Free PMC article. Review.
-
Heart-type fatty acid binding protein (H-FABP) as an early biomarker in sepsis-induced cardiomyopathy: a prospective observational study.Lipids Health Dis. 2024 Sep 4;23(1):283. doi: 10.1186/s12944-024-02264-0. Lipids Health Dis. 2024. PMID: 39232765 Free PMC article.
-
Probing the interaction of brain fatty acid binding protein (B-FABP) with model membranes.PLoS One. 2013;8(3):e60198. doi: 10.1371/journal.pone.0060198. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555925 Free PMC article.
-
Effect of methotrexate treatment on the expression of epidermal-fatty acid-binding protein (E-FABP) and apolipoproteins in patients with psoriasis.Postepy Dermatol Alergol. 2020 Jun;37(3):401-406. doi: 10.5114/ada.2020.96109. Epub 2020 Jul 16. Postepy Dermatol Alergol. 2020. PMID: 32792883 Free PMC article.
References
-
- Allen, G. W., J. W. Liu, and M. De Leon. 2000. Depletion of a fatty acid-binding protein impairs neurite outgrowth in PC12 cells. Brain Res. Mol. Brain Res. 76:315-324. - PubMed
-
- Banaszak, L., N. Winter, Z. Xu, D. A. Bernlohr, S. Cowan, and T. A. Jones. 1994. Lipid-binding proteins: a family of fatty acid and retinoid transport proteins. Adv. Protein Chem. 45:89-151. - PubMed
-
- Bass, N. M. 1990. Fatty acid-binding protein expression in the liver: its regulation and relationship to the zonation of fatty acid metabolism. Mol. Cell. Biochem. 98:167-176. - PubMed
-
- Budhu, A., R. Gillilan, and N. Noy. 2001. Localization of the RAR interaction domain of cellular retinoic acid binding protein-II. J. Mol. Biol. 305:939-949. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
