The lateral organ boundaries gene defines a novel, plant-specific gene family
- PMID: 12068116
- PMCID: PMC161698
- DOI: 10.1104/pp.010926
The lateral organ boundaries gene defines a novel, plant-specific gene family
Abstract
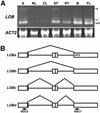
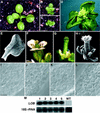
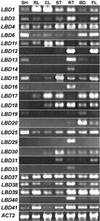
The LATERAL ORGAN BOUNDARIES (LOB) gene in Arabidopsis defines a new conserved protein domain. LOB is expressed in a band of cells at the adaxial base of all lateral organs formed from the shoot apical meristem and at the base of lateral roots. LOB encodes a predicted protein that does not have recognizable functional motifs, but that contains a conserved domain (the LOB domain) that is present in 42 other Arabidopsis proteins and in proteins from a variety of other plant species. Proteins showing similarity to the LOB domain were not found outside of plant databases, indicating that this unique protein may play a role in plant-specific processes. Genes encoding LOB domain proteins are expressed in a variety of temporal- and tissue-specific patterns, suggesting that they may function in diverse processes. Loss-of-function LOB mutants have no detectable phenotype under standard growth conditions, suggesting that LOB is functionally redundant or required during growth under specific environmental conditions. Ectopic expression of LOB leads to alterations in the size and shape of leaves and floral organs and causes male and female sterility. The expression of LOB at the base of lateral organs suggests a potential role for LOB in lateral organ development.
Figures



Similar articles
-
Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity.Plant Cell. 2003 Nov;15(11):2532-50. doi: 10.1105/tpc.014928. Epub 2003 Oct 10. Plant Cell. 2003. PMID: 14555694 Free PMC article.
-
Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members.Plant J. 2009 May;58(3):525-37. doi: 10.1111/j.1365-313X.2009.03797.x. Epub 2009 Jan 19. Plant J. 2009. PMID: 19154202 Free PMC article.
-
Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries.Proc Natl Acad Sci U S A. 2012 Dec 18;109(51):21146-51. doi: 10.1073/pnas.1210789109. Epub 2012 Dec 4. Proc Natl Acad Sci U S A. 2012. PMID: 23213252 Free PMC article.
-
KNAT6 gene of Arabidopsis is expressed in roots and is required for correct lateral root formation.Plant Mol Biol. 2004 Jan;54(1):71-84. doi: 10.1023/B:PLAN.0000028772.22892.2d. Plant Mol Biol. 2004. PMID: 15159635
-
ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis.Development. 2005 Mar;132(5):897-911. doi: 10.1242/dev.01642. Epub 2005 Jan 26. Development. 2005. PMID: 15673576
Cited by
-
Single-cell RNA-seq reveals a link of ovule abortion and sugar transport in Camellia oleifera.Front Plant Sci. 2024 Feb 2;15:1274013. doi: 10.3389/fpls.2024.1274013. eCollection 2024. Front Plant Sci. 2024. PMID: 38371413 Free PMC article.
-
Perspectives on leaf dorsoventral polarity.J Plant Res. 2010 May;123(3):281-90. doi: 10.1007/s10265-010-0336-3. Epub 2010 Apr 6. J Plant Res. 2010. PMID: 20369373 Review.
-
LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins.Nucleic Acids Res. 2007;35(19):6663-71. doi: 10.1093/nar/gkm775. Epub 2007 Oct 2. Nucleic Acids Res. 2007. PMID: 17913740 Free PMC article.
-
Plantacyanin plays a role in reproduction in Arabidopsis.Plant Physiol. 2005 Jun;138(2):778-89. doi: 10.1104/pp.105.063388. Epub 2005 May 20. Plant Physiol. 2005. PMID: 15908590 Free PMC article.
-
A PXY-Mediated Transcriptional Network Integrates Signaling Mechanisms to Control Vascular Development in Arabidopsis.Plant Cell. 2020 Feb;32(2):319-335. doi: 10.1105/tpc.19.00562. Epub 2019 Dec 5. Plant Cell. 2020. PMID: 31806676 Free PMC article.
References
-
- Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESSgenes. Development. 1999;126:1563–1570. - PubMed
-
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8actin subclass in vegetative tissues. Plant J. 1996;10:107–121. - PubMed
-
- Barton MK, Poethig RS. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemlessmutant. Development. 1993;119:823–831.
-
- Berleth T, Jürgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsisembryo. Development. 1993;118:575–587.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

