p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor
- PMID: 12032085
- PMCID: PMC126028
- DOI: 10.1093/emboj/21.11.2724
p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor
Abstract
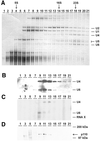
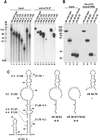
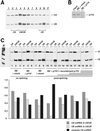
During each spliceosome cycle, the U6 snRNA undergoes extensive structural rearrangements, alternating between singular, U4-U6 and U6-U2 base-paired forms. In Saccharomyces cerevisiae, Prp24 functions as an snRNP recycling factor, reannealing U4 and U6 snRNAs. By database searching, we have identified a Prp24-related human protein previously described as p110(nrb) or SART3. p110 contains in its C-terminal region two RNA recognition motifs (RRMs). The N-terminal two-thirds of p110, for which there is no counterpart in the S.cerevisiae Prp24, carries seven tetratricopeptide repeat (TPR) domains. p110 homologs sharing the same domain structure also exist in several other eukaryotes. p110 is associated with the mammalian U6 and U4/U6 snRNPs, but not with U4/U5/U6 tri-snRNPs nor with spliceosomes. Recom binant p110 binds in vitro specifically to human U6 snRNA, requiring an internal U6 region. Using an in vitro recycling assay, we demonstrate that p110 functions in the reassembly of the U4/U6 snRNP. In summary, p110 represents the human ortholog of Prp24, and associates only transiently with U6 and U4/U6 snRNPs during the recycling phase of the spliceosome cycle.
Figures




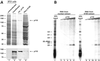
Similar articles
-
Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling.Mol Cell Biol. 2004 Sep;24(17):7392-401. doi: 10.1128/MCB.24.17.7392-7401.2004. Mol Cell Biol. 2004. PMID: 15314151 Free PMC article.
-
Recycling of the U12-type spliceosome requires p110, a component of the U6atac snRNP.Mol Cell Biol. 2004 Feb;24(4):1700-8. doi: 10.1128/MCB.24.4.1700-1708.2004. Mol Cell Biol. 2004. PMID: 14749385 Free PMC article.
-
Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell's and yeast spliceosomal U4/U6.U5 tri-snRNP particles.J Mol Biol. 2002 Apr 12;317(5):631-49. doi: 10.1006/jmbi.2002.5451. J Mol Biol. 2002. PMID: 11955014
-
CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast.Curr Opin Struct Biol. 2016 Feb;36:48-57. doi: 10.1016/j.sbi.2015.12.005. Epub 2016 Jan 21. Curr Opin Struct Biol. 2016. PMID: 26803803 Free PMC article. Review.
-
Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles.Curr Opin Struct Biol. 1999 Apr;9(2):222-30. doi: 10.1016/s0959-440x(99)80032-3. Curr Opin Struct Biol. 1999. PMID: 10322216 Review.
Cited by
-
Analysis of copy number alterations reveals the lncRNA ALAL-1 as a regulator of lung cancer immune evasion.J Cell Biol. 2020 Sep 7;219(9):e201908078. doi: 10.1083/jcb.201908078. J Cell Biol. 2020. PMID: 32858747 Free PMC article.
-
An RNA electrophoretic mobility shift and mutational analysis of rnp-4f 5'-UTR intron splicing regulatory proteins in Drosophila reveals a novel new role for a dADAR protein isoform.Gene. 2012 Dec 15;511(2):161-8. doi: 10.1016/j.gene.2012.09.088. Epub 2012 Sep 28. Gene. 2012. PMID: 23026215 Free PMC article.
-
Long noncoding RNA ABHD11-AS1 interacts with SART3 and regulates CD44 RNA alternative splicing to promote lung carcinogenesis.Environ Int. 2024 Mar;185:108494. doi: 10.1016/j.envint.2024.108494. Epub 2024 Feb 10. Environ Int. 2024. PMID: 38364571 Free PMC article.
-
Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling.Mol Cell Biol. 2004 Sep;24(17):7392-401. doi: 10.1128/MCB.24.17.7392-7401.2004. Mol Cell Biol. 2004. PMID: 15314151 Free PMC article.
-
The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination.J Biol Chem. 2014 Mar 28;289(13):8916-30. doi: 10.1074/jbc.M114.551754. Epub 2014 Feb 13. J Biol Chem. 2014. PMID: 24526689 Free PMC article.
References
-
- Blatch G.L. and Lassle,M. (1999) The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. BioEssays, 21, 932–939. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

