Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis
- PMID: 12021352
- PMCID: PMC136188
- DOI: 10.1128/jvi.76.12.6185-6196.2002
Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis
Abstract
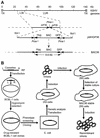
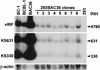
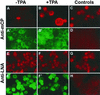
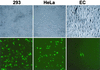
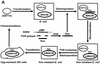
Kaposi's sarcoma-associated herpesvirus (KSHV) is etiologically associated with Kaposi's sarcoma and several other malignancies. The lack of an efficient infection system has impeded the understanding of KSHV-related pathogenesis. A genetic approach was used to isolate infectious KSHV. Recombinant bacteria artificial chromosome (BAC) KSHV containing hygromycin resistance and green fluorescent protein (GFP) markers was generated by homologous recombination in KSHV-infected BCBL-1 cells. Recombinant KSHV genomes from cell clones that were resistant to hygromycin, expressed GFP, and produced infectious virions after induction with tetradecanoyl phorbol acetate (TPA) were rescued in Escherichia coli and reconstituted in 293 cells. Several 293 cell lines resulting from infection with recombinant virions induced from a full-length recombinant KSHV genome, named BAC36, were obtained. BAC36 virions established stable latent infection in 293 cells, harboring 1 to 2 copies of viral genome per cell and expressing viral latent proteins, with approximately 0.5% of cells undergoing spontaneous lytic replication, which is reminiscent of KSHV infection in Kaposi's sarcoma tumors. TPA treatment induced BAC36-infected 293 cell lines into productive lytic replication, expressing lytic proteins and producing virions that efficiently infected normal 293 cells with a approximately 50% primary infection rate. BAC36 virions were also infectious to HeLa and E6E7-immortalized human endothelial cells. Since BAC36 can be efficiently shuttled between bacteria and mammalian cells, it is useful for KSHV genetic analysis. The feasibility of the system was illustrated through the generation of a KSHV mutant with the vIRF gene deleted. This cellular model is useful for the investigation of KSHV infection and pathogenesis.
Figures


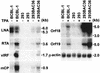

Similar articles
-
Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells.J Virol. 2015 Mar;89(6):3093-111. doi: 10.1128/JVI.02507-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552714 Free PMC article.
-
De novo infection and serial transmission of Kaposi's sarcoma-associated herpesvirus in cultured endothelial cells.J Virol. 2002 Mar;76(5):2440-8. doi: 10.1128/jvi.76.5.2440-2448.2002. J Virol. 2002. PMID: 11836422 Free PMC article.
-
Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures.J Virol. 2001 Jun;75(12):5614-26. doi: 10.1128/JVI.75.12.5614-5626.2001. J Virol. 2001. PMID: 11356969 Free PMC article.
-
Pathological Features of Kaposi's Sarcoma-Associated Herpesvirus Infection.Adv Exp Med Biol. 2018;1045:357-376. doi: 10.1007/978-981-10-7230-7_16. Adv Exp Med Biol. 2018. PMID: 29896675 Review.
-
[Replication Machinery of Kaposi's Sarcoma-associated Herpesvirus and Drug Discovery Research].Yakugaku Zasshi. 2019;139(1):69-73. doi: 10.1248/yakushi.18-00164-2. Yakugaku Zasshi. 2019. PMID: 30606932 Review. Japanese.
Cited by
-
Leucine zipper domain is required for Kaposi sarcoma-associated herpesvirus (KSHV) K-bZIP protein to interact with histone deacetylase and is important for KSHV replication.J Biol Chem. 2012 May 4;287(19):15622-34. doi: 10.1074/jbc.M111.315861. Epub 2012 Mar 13. J Biol Chem. 2012. PMID: 22416134 Free PMC article.
-
Human cytomegalovirus: bacterial artificial chromosome (BAC) cloning and genetic manipulation.Curr Protoc Microbiol. 2012 Feb;Chapter 14:Unit14E.4. doi: 10.1002/9780471729259.mc14e04s24. Curr Protoc Microbiol. 2012. PMID: 22307551 Free PMC article.
-
Envelope glycoprotein gB of Kaposi's sarcoma-associated herpesvirus is essential for egress from infected cells.J Virol. 2005 Sep;79(17):10952-67. doi: 10.1128/JVI.79.17.10952-10967.2005. J Virol. 2005. PMID: 16103147 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways.J Virol. 2005 Dec;79(24):15027-37. doi: 10.1128/JVI.79.24.15027-15037.2005. J Virol. 2005. PMID: 16306573 Free PMC article.
-
Herpesvirus BACs: past, present, and future.J Biomed Biotechnol. 2011;2011:124595. doi: 10.1155/2011/124595. Epub 2010 Oct 27. J Biomed Biotechnol. 2011. PMID: 21048927 Free PMC article. Review.
References
-
- Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. - PubMed
-
- Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. D. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpes virus (KSHV) infects endothelial and spindle cells. Nat. Med. 1:1274-1278. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

