Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4
- PMID: 11997514
- PMCID: PMC133820
- DOI: 10.1128/MCB.22.11.3794-3802.2002
Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4
Abstract
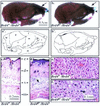
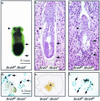
In a gene trap screen we recovered a mouse mutant line in which an insertion generated a null allele of the Brd4 gene. Brd4 belongs to the Fsh/Brd family, a group of structurally related proteins characterized by the association of two bromodomains and one extraterminal domain. Members of this family include Brd2/Ring3/Fsrg1 in mammals, fs(1)h in Drosophila, and Bdf1 in Saccharomyces cerevisiae. Brd4 heterozygotes display pre- and postnatal growth defects associated with a reduced proliferation rate. These mice also exhibit a variety of anatomical abnormalities: head malformations, absence of subcutaneous fat, cataracts, and abnormal liver cells. In primary cell cultures, heterozygous cells also display reduced proliferation rates and moderate sensitivity to methyl methanesulfonate. Embryos nullizygous for Brd4 die shortly after implantation and are compromised in their ability to maintain an inner cell mass in vitro, suggesting a role in fundamental cellular processes. Finally, sequence comparisons suggest that Brd4 is likely to correspond to the Brd-like element of the mediator of transcriptional regulation isolated by Y. W. Jiang, P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg (Proc. Natl. Acad. Sci. USA 95:8538-8543, 1998) and the Brd4 mutant phenotype is discussed in light of this result. Together, our results provide the first genetic evidence for an in vivo role in mammals for a member of the Fsh/Brd family.
Figures




Similar articles
-
The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation.J Biol Chem. 2007 May 4;282(18):13141-5. doi: 10.1074/jbc.R700001200. Epub 2007 Feb 28. J Biol Chem. 2007. PMID: 17329240 Review.
-
Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest.J Virol. 2006 Nov;80(21):10772-86. doi: 10.1128/JVI.00804-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928766 Free PMC article.
-
BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells.Oncogene. 2008 Apr 3;27(15):2237-42. doi: 10.1038/sj.onc.1210852. Epub 2007 Oct 15. Oncogene. 2008. PMID: 17934517
-
Brd4 links chromatin targeting to HPV transcriptional silencing.Genes Dev. 2006 Sep 1;20(17):2383-96. doi: 10.1101/gad.1448206. Epub 2006 Aug 18. Genes Dev. 2006. PMID: 16921027 Free PMC article.
-
Bromodomain 4: a cellular Swiss army knife.J Leukoc Biol. 2016 Oct;100(4):679-686. doi: 10.1189/jlb.2RI0616-250R. Epub 2016 Jul 22. J Leukoc Biol. 2016. PMID: 27450555 Free PMC article. Review.
Cited by
-
When are the BET factors the most sensitive to bromodomain inhibitors?Transcription. 2013 Mar-Apr;4(2):54-7. doi: 10.4161/trns.23191. Epub 2013 Feb 14. Transcription. 2013. PMID: 23412362 Free PMC article.
-
Brd4-independent transcriptional repression function of the papillomavirus e2 proteins.J Virol. 2007 Sep;81(18):9612-22. doi: 10.1128/JVI.00447-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626100 Free PMC article.
-
The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.J Biol Chem. 2018 May 11;293(19):7160-7175. doi: 10.1074/jbc.RA117.000678. Epub 2018 Mar 22. J Biol Chem. 2018. PMID: 29567837 Free PMC article.
-
BRD4 as a Therapeutic Target in Pulmonary Diseases.Int J Mol Sci. 2023 Aug 25;24(17):13231. doi: 10.3390/ijms241713231. Int J Mol Sci. 2023. PMID: 37686037 Free PMC article. Review.
-
The double bromodomain protein Brd2 promotes B cell expansion and mitogenesis.J Leukoc Biol. 2014 Mar;95(3):451-60. doi: 10.1189/jlb.1112588. Epub 2013 Dec 6. J Leukoc Biol. 2014. PMID: 24319289 Free PMC article.
References
-
- Beck, S., I. Hanson, A. Kelly, D. J. Pappin, and J. Trowsdale. 1992. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Sequence 2:203-210. - PubMed
-
- Bergmann, E., and J. M. Egly. 2001. Trichothiodystrophy, a transcription syndrome. Trends Genet. 17:279-286. - PubMed
-
- Berneburg, M., and A. R. Lehmann. 2001. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet. 43:71-102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
