Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase
- PMID: 11971140
- PMCID: PMC150687
- DOI: 10.1105/tpc.010480
Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase
Abstract
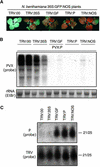
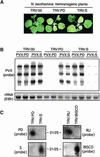
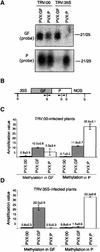
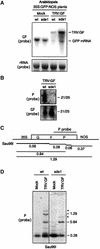
RNA silencing is a sequence-specific RNA degradation process that follows the recognition of double-stranded RNA. Here, we show that virus vectors carrying parts of a green fluorescent protein (GFP) transgene targeted RNA silencing in Nicotiana benthamiana and Arabidopsis against the entire GFP RNA. These results indicate that there was spreading of RNA targeting from the initiator region into the adjacent 5' and 3' regions of the target gene. Spreading was accompanied by methylation of the corresponding GFP DNA. It also was dependent on transcription of the transgene and on the putative RNA-dependent RNA polymerase, SDE1/SGS2. These findings indicate that SDE1/SGS2 produces double-stranded RNA using the target RNA as a template. RNA silencing of ribulose-1,5-bisphosphate carboxylase/oxygenase and phytoene desaturase was not associated with the spreading of RNA targeting or DNA methylation, indicating that these endogenous RNAs are not templates for SDE1/SGS2.
Figures
Similar articles
-
RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing.Plant Cell. 1999 Dec;11(12):2291-301. doi: 10.1105/tpc.11.12.2291. Plant Cell. 1999. PMID: 10590159 Free PMC article.
-
Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants.Plant J. 2004 Jun;38(6):1004-14. doi: 10.1111/j.1365-313X.2004.02103.x. Plant J. 2004. PMID: 15165191
-
Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector.Plant J. 2001 Feb;25(4):417-25. doi: 10.1046/j.1365-313x.2001.00976.x. Plant J. 2001. PMID: 11260498
-
A conserved mechanism for post-transcriptional gene silencing?Genome Biol. 2000;1(3):REVIEWS1018. doi: 10.1186/gb-2000-1-3-reviews1018. Epub 2000 Sep 13. Genome Biol. 2000. PMID: 11178243 Free PMC article. Review.
-
RNA-based silencing strategies in plants.Curr Opin Genet Dev. 2001 Apr;11(2):221-7. doi: 10.1016/s0959-437x(00)00183-0. Curr Opin Genet Dev. 2001. PMID: 11250148 Review.
Cited by
-
Primary and secondary siRNA synthesis triggered by RNAs from food bacteria in the ciliate Paramecium tetraurelia.Nucleic Acids Res. 2015 Feb 18;43(3):1818-33. doi: 10.1093/nar/gku1331. Epub 2015 Jan 15. Nucleic Acids Res. 2015. PMID: 25593325 Free PMC article.
-
Primary and secondary siRNAs in geminivirus-induced gene silencing.PLoS Pathog. 2012 Sep;8(9):e1002941. doi: 10.1371/journal.ppat.1002941. Epub 2012 Sep 27. PLoS Pathog. 2012. PMID: 23028332 Free PMC article.
-
Molecular cloning and characterization of an inducible RNA-dependent RNA polymerase gene, GhRdRP, from cotton (Gossypium hirsutum L.).Mol Biol Rep. 2009 Jan;36(1):47-56. doi: 10.1007/s11033-007-9150-y. Epub 2007 Oct 11. Mol Biol Rep. 2009. PMID: 17929195
-
SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene.Plant Physiol. 2003 Nov;133(3):1397-406. doi: 10.1104/pp.103.028019. Epub 2003 Oct 9. Plant Physiol. 2003. PMID: 14551330 Free PMC article.
-
A 22-nt artificial microRNA mediates widespread RNA silencing in Arabidopsis.Plant J. 2013 Nov;76(3):519-29. doi: 10.1111/tpj.12306. Epub 2013 Oct 3. Plant J. 2013. PMID: 23937661 Free PMC article.
References
-
- Al-Kaff, N.S., Covey, S.N., Kreike, M.M., Page, A.M., Pinder, R., and Dale, P.J. (1998). Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279, 2113–2115. - PubMed
-
- Aravin, A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027. - PubMed
-
- Baulcombe, D.C. (1999). Gene silencing: RNA makes RNA makes no protein. Curr. Biol. 9, R599–R601. - PubMed
-
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

