Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus
- PMID: 11967315
- PMCID: PMC136159
- DOI: 10.1128/jvi.76.10.4987-4999.2002
Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus
Abstract
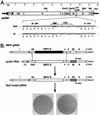
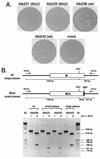
The coronavirus membrane (M) protein is the most abundant virion protein and the key component in viral assembly and morphogenesis. The M protein of mouse hepatitis virus (MHV) is an integral membrane protein with a short ectodomain, three transmembrane segments, and a large carboxy-terminal endodomain facing the interior of the viral envelope. The carboxy terminus of MHV M has previously been shown to be extremely sensitive to mutation, both in a virus-like particle expression system and in the intact virion. We have constructed a mutant, M(Delta)2, containing a two-amino-acid truncation of the M protein that was previously thought to be lethal. This mutant was isolated by means of targeted RNA recombination with a powerful host range-based selection allowed by the interspecies chimeric virus fMHV (MHV containing the ectodomain of the feline infectious peritonitis virus S protein). Analysis of multiple second-site revertants of the M(Delta)2 mutant has revealed changes in regions of both the M protein and the nucleocapsid (N) protein that can compensate for the loss of the last two residues of the M protein. Our data thus provide the first genetic evidence for a structural interaction between the carboxy termini of the M and N proteins of MHV. In addition, this work demonstrates the efficacy of targeted recombination with fMHV for the systematic genetic analysis of coronavirus structural protein interactions.
Figures





Similar articles
-
A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein.J Virol. 2005 Nov;79(21):13285-97. doi: 10.1128/JVI.79.21.13285-13297.2005. J Virol. 2005. PMID: 16227251 Free PMC article.
-
Analyses of Coronavirus Assembly Interactions with Interspecies Membrane and Nucleocapsid Protein Chimeras.J Virol. 2016 Apr 14;90(9):4357-4368. doi: 10.1128/JVI.03212-15. Print 2016 May. J Virol. 2016. PMID: 26889024 Free PMC article.
-
Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier.J Virol. 2000 Feb;74(3):1393-406. doi: 10.1128/jvi.74.3.1393-1406.2000. J Virol. 2000. PMID: 10627550 Free PMC article.
-
The Hepatitis B Virus Envelope Proteins: Molecular Gymnastics Throughout the Viral Life Cycle.Annu Rev Virol. 2020 Sep 29;7(1):263-288. doi: 10.1146/annurev-virology-092818-015508. Epub 2020 Jun 29. Annu Rev Virol. 2020. PMID: 32600157 Review.
-
Coronavirus genomic RNA packaging.Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review.
Cited by
-
Coronavirus M Protein Trafficking in Epithelial Cells Utilizes a Myosin Vb Splice Variant and Rab10.Cells. 2024 Jan 10;13(2):126. doi: 10.3390/cells13020126. Cells. 2024. PMID: 38247817 Free PMC article.
-
Covid-19: virology, variants, and vaccines.BMJ Med. 2022 Apr 1;1(1):e000040. doi: 10.1136/bmjmed-2021-000040. eCollection 2022. BMJ Med. 2022. PMID: 36936563 Free PMC article. Review.
-
Structural insights into ribonucleoprotein dissociation by nucleocapsid protein interacting with non-structural protein 3 in SARS-CoV-2.Commun Biol. 2023 Feb 18;6(1):193. doi: 10.1038/s42003-023-04570-2. Commun Biol. 2023. PMID: 36806252 Free PMC article.
-
Biophysical Modeling of SARS-CoV-2 Assembly: Genome Condensation and Budding.Viruses. 2022 Sep 20;14(10):2089. doi: 10.3390/v14102089. Viruses. 2022. PMID: 36298645 Free PMC article.
-
Antiviral effects of Korean Red Ginseng on human coronavirus OC43.J Ginseng Res. 2023 Mar;47(2):329-336. doi: 10.1016/j.jgr.2022.09.009. Epub 2022 Oct 5. J Ginseng Res. 2023. PMID: 36217314 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

