Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism
- PMID: 11932416
- PMCID: PMC155063
- DOI: 10.1128/jvi.76.9.4507-4519.2002
Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism
Abstract
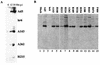
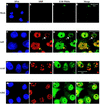
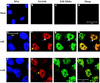
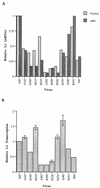
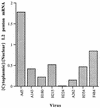
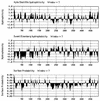
The human subgroup C adenoviral E1B 55-kDa protein cooperates with the viral E4 Orf6 protein to induce selective export of viral, late mRNAs from the nucleus to the cytoplasm. Previous studies have suggested that such preferential transport of viral mRNA and the concomitant inhibition of export of cellular mRNAs are the result of viral colonization of specialized microenvironments within the nucleus. However, neither the molecular basis of this phenomenon nor the mechanism by which the E1B 55-kDa protein acts has been elucidated. We therefore examined viral late mRNA metabolism in HeLa cells infected with a series of mutant viruses that carry insertions at various positions in the E1B protein coding sequence (P. R. Yew, C. C. Kao, and A. J. Berk, Virology 179:795-805, 1990). All the mutations examined impaired cytoplasmic accumulation of viral L2 mRNAs and reduced L2 mRNA export efficiency. However, in most cases these defects could be ascribed to reduced E1B 55-kDa protein concentration or the unexpected failure of the altered E1B proteins to enter the nucleus efficiently. The latter property, the pleiotropic defects associated with all the mutations that impaired nuclear entry of the E1B protein, and consideration of its primary sequence suggest that these insertions result in misfolding of the protein. Insertion of four amino acids at residue 143 also inhibited viral mRNA export but resulted in increased rather than decreased accumulation of the E1B 55-kDa protein in the nucleus. This mutation specifically impaired the previously described association of the E1B protein with intranuclear structures that correspond to sites of adenoviral DNA replication and transcription (D. Ornelles and T. Shenk, J. Virol. 65:424-439, 1991) and the colocalization of the E1B and E4 Orf6 proteins. As this insertion has been shown to inhibit the interaction of the E1B with the E4 Orf6 protein in infected cell extracts (S. Rubenwolf, H. Schütt, M. Nevels, H. Wolf, and T. Dobner, J. Virol. 71:1115-1123, 1997), these phenotypes provide direct support for the hypothesis that selective viral mRNA export is determined by the functional organization of the infected cell nucleus.
Figures


Similar articles
-
Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication.J Virol. 1999 Sep;73(9):7474-88. doi: 10.1128/JVI.73.9.7474-7488.1999. J Virol. 1999. PMID: 10438837 Free PMC article.
-
Export of adenoviral late mRNA from the nucleus requires the Nxf1/Tap export receptor.J Virol. 2011 Feb;85(4):1429-38. doi: 10.1128/JVI.02108-10. Epub 2010 Dec 1. J Virol. 2011. PMID: 21123381 Free PMC article.
-
Adenovirus E1B 55-kilodalton protein is required for both regulation of mRNA export and efficient entry into the late phase of infection in normal human fibroblasts.J Virol. 2006 Jan;80(2):964-74. doi: 10.1128/JVI.80.2.964-974.2006. J Virol. 2006. PMID: 16378998 Free PMC article.
-
Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins.Curr Top Microbiol Immunol. 2003;272:287-330. doi: 10.1007/978-3-662-05597-7_10. Curr Top Microbiol Immunol. 2003. PMID: 12747554 Review.
-
Release of viruses and viral DNA from nucleus to cytoplasm of HeLa cells at late stages of productive adenovirus infection as revealed by electron microscope in situ hybridization.Biol Cell. 1998 Jan;90(1):5-38. doi: 10.1016/s0248-4900(98)80230-x. Biol Cell. 1998. PMID: 9691424 Review.
Cited by
-
Human adenovirus oncolytic properties and the inhibitory role of E4 orf4 and E4 orf6/7 on endogenously activated NF-κB.Biochem Biophys Rep. 2023 Dec 22;37:101616. doi: 10.1016/j.bbrep.2023.101616. eCollection 2024 Mar. Biochem Biophys Rep. 2023. PMID: 38205184 Free PMC article.
-
Adenovirus E1B 55-kilodalton oncoprotein binds to Daxx and eliminates enhancement of p53-dependent transcription by Daxx.J Virol. 2003 Nov;77(21):11809-21. doi: 10.1128/jvi.77.21.11809-11821.2003. J Virol. 2003. PMID: 14557665 Free PMC article.
-
Going viral: a review of replication-selective oncolytic adenoviruses.Oncotarget. 2015 Aug 21;6(24):19976-89. doi: 10.18632/oncotarget.5116. Oncotarget. 2015. PMID: 26280277 Free PMC article. Review.
-
The human adenovirus type 5 E1B 55kDa protein interacts with RNA promoting timely DNA replication and viral late mRNA metabolism.PLoS One. 2019 Apr 3;14(4):e0214882. doi: 10.1371/journal.pone.0214882. eCollection 2019. PLoS One. 2019. PMID: 30943256 Free PMC article.
-
RUNX1 permits E4orf6-directed nuclear localization of the adenovirus E1B-55K protein and associates with centers of viral DNA and RNA synthesis.J Virol. 2008 Jul;82(13):6395-408. doi: 10.1128/JVI.00043-08. Epub 2008 Apr 16. J Virol. 2008. PMID: 18417565 Free PMC article.
References
-
- Aspegren, A., C. Rabino, and E. Bridge. 1998. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp. Cell Res. 245:203-213. - PubMed
-
- Bachi, A., I. C. Braun, J. P. Rodrigues, N. Pante, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Gorlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. - PMC - PubMed
-
- Baeuerle, P. A., and D. Baltimore. 1996. NF-κB: ten years after. Cell 87:13-20. - PubMed
-
- Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

