Probability-based protein secondary structure identification using combined NMR chemical-shift data
- PMID: 11910028
- PMCID: PMC2373532
- DOI: 10.1110/ps.3180102
Probability-based protein secondary structure identification using combined NMR chemical-shift data
Abstract
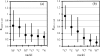
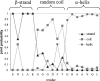
For a long time, NMR chemical shifts have been used to identify protein secondary structures. Currently, this is accomplished through comparing the observed (1)H(alpha), (13)C(alpha), (13)C(beta), or (13)C' chemical shifts with the random coil values. Here, we present a new protocol, which is based on the joint probability of each of the three secondary structural types (beta-strand, alpha-helix, and random coil) derived from chemical-shift data, to identify the secondary structure. In combination with empirical smooth filters/functions, this protocol shows significant improvements in the accuracy and the confidence of identification. Updated chemical-shift statistics are reported, on the basis of which the reliability of using chemical shift to identify protein secondary structure is evaluated for each nucleus. The reliability varies greatly among the 20 amino acids, but, on average, is in the order of: (13)C(alpha)>(13)C'>(1)H(alpha)>(13)C(beta)>(15)N>(1)H(N) to distinguish an alpha-helix from a random coil; and (1)H(alpha)>(13)C(beta) >(1)H(N) approximately (13)C(alpha) approximately (13)C' approximately (15)N for a beta-strand from a random coil. Amide (15)N and (1)H(N) chemical shifts, which are generally excluded from the application, in fact, were found to be helpful in distinguishing a beta-strand from a random coil. In addition, the chemical-shift statistical data are compared with those reported previously, and the results are discussed. A JAVA User Interface program has been developed to make the entire procedure fully automated and is available via http://ccsr3150-p3.stanford.edu.
Figures
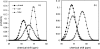
Similar articles
-
Uncovering symmetry-breaking vector and reliability order for assigning secondary structures of proteins from atomic NMR chemical shifts in amino acids.J Biomol NMR. 2011 Dec;51(4):411-24. doi: 10.1007/s10858-011-9579-0. Epub 2011 Oct 30. J Biomol NMR. 2011. PMID: 22038647
-
Comparison between the phi distribution of the amino acids in the protein database and NMR data indicates that amino acids have various phi propensities in the random coil conformation.J Mol Biol. 1995 Nov 24;254(2):322-33. doi: 10.1006/jmbi.1995.0619. J Mol Biol. 1995. PMID: 7490751
-
Protein chemical shifts arising from alpha-helices and beta-sheets depend on solvent exposure.Proc Natl Acad Sci U S A. 2004 Dec 14;101(50):17394-7. doi: 10.1073/pnas.0407969101. Epub 2004 Dec 1. Proc Natl Acad Sci U S A. 2004. PMID: 15574491 Free PMC article.
-
Protein chemical shift analysis: a practical guide.Biochem Cell Biol. 1998;76(2-3):153-63. doi: 10.1139/bcb-76-2-3-153. Biochem Cell Biol. 1998. PMID: 9923684 Review.
-
Two-dimensional NMR spectroscopy: an application to the study of flexibility of protein molecules.Adv Biophys. 1981;14:139-204. Adv Biophys. 1981. PMID: 7015809 Review.
Cited by
-
Deuterium isotope shifts for backbone ¹H, ¹⁵N and ¹³C nuclei in intrinsically disordered protein α-synuclein.J Biomol NMR. 2012 Oct;54(2):181-91. doi: 10.1007/s10858-012-9666-x. Epub 2012 Sep 8. J Biomol NMR. 2012. PMID: 22960996 Free PMC article.
-
The effects of threonine phosphorylation on the stability and dynamics of the central molecular switch region of 18.5-kDa myelin basic protein.PLoS One. 2013 Jul 5;8(7):e68175. doi: 10.1371/journal.pone.0068175. Print 2013. PLoS One. 2013. PMID: 23861868 Free PMC article.
-
EFG-CS: Predicting chemical shifts from amino acid sequences with protein structure prediction using machine learning and deep learning models.Protein Sci. 2024 Aug;33(8):e5096. doi: 10.1002/pro.5096. Protein Sci. 2024. PMID: 38979954 Free PMC article.
-
Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation.Protein Sci. 2006 Dec;15(12):2795-804. doi: 10.1110/ps.062465306. Epub 2006 Nov 6. Protein Sci. 2006. PMID: 17088319 Free PMC article.
-
Structural conservation of HBV-like capsid proteins over hundreds of millions of years despite the shift from non-enveloped to enveloped life-style.Nat Commun. 2023 Mar 22;14(1):1574. doi: 10.1038/s41467-023-37068-w. Nat Commun. 2023. PMID: 36949039 Free PMC article.
References
-
- Colloch, N., Etchebest, C., Thoreau, E., Henrissat, B, and Mornon, J.P. 1993. Comparison of three algorithms for the assignment of secondary structure in proteins: The advantages of a consensus assignment. Protein Eng. 377–382. - PubMed
-
- Cuff, J.A and Barton, G.J. 1999. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins: Struct. Funct. Genet. 34 508–519. - PubMed
-
- Frishman, D and Argos, P. 1995. Knowledge-based protein secondary structure assignment. Proteins 23 566–579. - PubMed
-
- Goto, N.K and Kay, L.E. 2000. New developments in isotope labeling strategies for protein solution NMR spectroscopy. Curr. Opin. Struct. Biol. 10 585–592. - PubMed
-
- Kabsch, W, and Sander, C. 1983. A dictionary of protein secondary structure. Biopolymers 22 2577–2637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

