Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology
- PMID: 11907260
- PMCID: PMC99597
- DOI: 10.1091/mbc.01-05-0275
Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology
Abstract
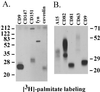
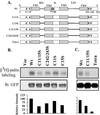
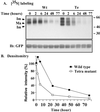
Here we demonstrate that multiple tetraspanin (transmembrane 4 superfamily) proteins are palmitoylated, in either the Golgi or a post-Golgi compartment. Using CD151 as a model tetraspanin, we identified and mutated intracellular N-terminal and C-terminal cysteine palmitoylation sites. Simultaneous mutations of C11, C15, C242, and C243 (each to serine) eliminated >90% of CD151 palmitoylation. Notably, palmitoylation had minimal influence on the density of tetraspanin protein complexes, did not promote tetraspanin localization into detergent-resistant microdomains, and was not required for CD151-alpha 3 beta 1 integrin association. However, the CD151 tetra mutant showed markedly diminished associations with other cell surface proteins, including other transmembrane 4 superfamily proteins (CD9, CD63). Thus, palmitoylation may be critical for assembly of the large network of cell surface tetraspanin-protein interactions, sometimes called the "tetraspanin web." Also, compared with wild-type CD151, the tetra mutant was much more diffusely distributed and showed markedly diminished stability during biosynthesis. Finally, expression of the tetra-CD151 mutant profoundly altered alpha 3 integrin-deficient kidney epithelial cells, such that they converted from a dispersed, elongated morphology to an epithelium-like cobblestone clustering. These results point to novel biochemical and biological functions for tetraspanin palmitoylation.
Figures

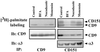






Similar articles
-
Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling.J Biol Chem. 2002 Oct 4;277(40):36991-7000. doi: 10.1074/jbc.M205265200. Epub 2002 Jul 10. J Biol Chem. 2002. PMID: 12110679
-
Analysis of the CD151-alpha3beta1 integrin and CD151-tetraspanin interactions by mutagenesis.J Biol Chem. 2001 Nov 2;276(44):41165-74. doi: 10.1074/jbc.M104041200. Epub 2001 Jul 30. J Biol Chem. 2001. PMID: 11479292
-
Palmitoylation supports assembly and function of integrin-tetraspanin complexes.J Cell Biol. 2004 Dec 20;167(6):1231-40. doi: 10.1083/jcb.200404100. J Cell Biol. 2004. PMID: 15611341 Free PMC article.
-
Tetraspanin protein contributions to cancer.Biochem Soc Trans. 2011 Apr;39(2):547-52. doi: 10.1042/BST0390547. Biochem Soc Trans. 2011. PMID: 21428937 Review.
-
Functional domains in tetraspanin proteins.Trends Biochem Sci. 2003 Feb;28(2):106-12. doi: 10.1016/S0968-0004(02)00014-2. Trends Biochem Sci. 2003. PMID: 12575999 Review.
Cited by
-
Tetraspanins as therapeutic targets in hematological malignancy: a concise review.Front Physiol. 2015 Mar 23;6:91. doi: 10.3389/fphys.2015.00091. eCollection 2015. Front Physiol. 2015. PMID: 25852576 Free PMC article. Review.
-
Molecular Regulation and Oncogenic Functions of TSPAN8.Cells. 2024 Jan 19;13(2):193. doi: 10.3390/cells13020193. Cells. 2024. PMID: 38275818 Free PMC article. Review.
-
Pathways and control of connexin oligomerization.Trends Cell Biol. 2006 Mar;16(3):159-66. doi: 10.1016/j.tcb.2006.01.006. Epub 2006 Feb 21. Trends Cell Biol. 2006. PMID: 16490353 Free PMC article. Review.
-
Effect of 2-fluoropalmitate, cerulenin and tunicamycin on the palmitoylation and intracellular translocation of myelin proteolipid protein.Neurochem Res. 2002 Dec;27(12):1669-75. doi: 10.1023/a:1021643229028. Neurochem Res. 2002. PMID: 12515321
-
alpha3beta1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion.J Cell Biol. 2003 Dec 22;163(6):1351-62. doi: 10.1083/jcb.200306067. J Cell Biol. 2003. PMID: 14691142 Free PMC article.
References
-
- Azorsa DO, Moog S, Cazenave JP, Lanza F. A general approach to the generation of monoclonal antibodies against members of the tetraspanin superfamily using recombinant GST fusion proteins. J Immunol Methods. 1999;229:35–48. - PubMed
-
- Barylko B, Gerber SH, Binns DD, Grichine N, Khvotchev M, Sudhof TC, Albanesi JP. A novel family of phos-phatidylinositol 4-kinases conserved from yeast to humans. J Biol Chem. 2001;276:7705–7708. - PubMed
-
- Berditchevski F, Bazzoni G, Hemler ME. Specific association of CD63 with the VLA-3 and VLA-6 integrins. J Biol Chem. 1995;270:17784–17790. - PubMed
-
- Berditchevski F, Chang S, Bodorova J, Hemler ME. Generation of monoclonal antibodies to integrin-associated proteins: evidence that α3β1 complexes with EMMPRIN/basigin/OX47/M6. J Biol Chem. 1997;272:29174–29180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

