Notch receptor cleavage depends on but is not directly executed by presenilins
- PMID: 11891288
- PMCID: PMC122640
- DOI: 10.1073/pnas.052017699
Notch receptor cleavage depends on but is not directly executed by presenilins
Abstract
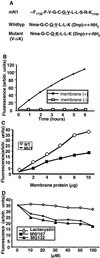
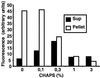
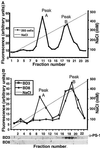
Notch receptors undergo three distinct proteolytic cleavages during maturation and activation. The third cleavage occurs within the plasma membrane and results in the release and translocation of the intracellular domain into the nucleus to execute Notch signaling. This so-called gamma-secretase cleavage is under the control of presenilins, but it is not known whether presenilins themselves carry out the cleavage or whether they act by means of yet-unidentified gamma-secretase(s). In this article, we show that Notch intracellular cleavage in intact cells completely depends on presenilins. In contrast, partial purification of the Notch cleavage activity reveals an activity, which is present only in protein extracts from presenilin-containing cells, and which does not comigrate with presenilin. This finding provides evidence for the existence of a specific Notch-processing activity, which is physically distinct from presenilins. We conclude from these experiments that presenilins are critically required for Notch intracellular cleavage but are not themselves directly mediating the cleavage.
Figures

Similar articles
-
Presenilins are required for gamma-secretase cleavage of beta-APP and transmembrane cleavage of Notch-1.Nat Cell Biol. 2000 Jul;2(7):463-5. doi: 10.1038/35017108. Nat Cell Biol. 2000. PMID: 10878814 No abstract available.
-
A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain.Nature. 1999 Apr 8;398(6727):518-22. doi: 10.1038/19083. Nature. 1999. PMID: 10206645
-
Notch oncoproteins depend on gamma-secretase/presenilin activity for processing and function.J Biol Chem. 2004 Jul 16;279(29):30771-80. doi: 10.1074/jbc.M309252200. Epub 2004 May 3. J Biol Chem. 2004. PMID: 15123653
-
Implication of APP secretases in notch signaling.J Mol Neurosci. 2001 Oct;17(2):171-81. doi: 10.1385/JMN:17:2:171. J Mol Neurosci. 2001. PMID: 11816790 Review.
-
The presenilins in Alzheimer's disease--proteolysis holds the key.Science. 1999 Oct 29;286(5441):916-9. doi: 10.1126/science.286.5441.916. Science. 1999. PMID: 10542139 Review.
Cited by
-
The large hydrophilic loop of presenilin 1 is important for regulating gamma-secretase complex assembly and dictating the amyloid beta peptide (Abeta) Profile without affecting Notch processing.J Biol Chem. 2010 Mar 19;285(12):8527-36. doi: 10.1074/jbc.M109.055590. Epub 2010 Jan 27. J Biol Chem. 2010. PMID: 20106965 Free PMC article.
-
Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor.J Cell Biol. 2004 Jul 5;166(1):73-83. doi: 10.1083/jcb.200310098. J Cell Biol. 2004. PMID: 15240571 Free PMC article.
-
Inhibition of gamma-secretase in Notch1 signaling pathway as a novel treatment for ovarian cancer.Oncotarget. 2017 Jan 31;8(5):8215-8225. doi: 10.18632/oncotarget.14152. Oncotarget. 2017. PMID: 28030808 Free PMC article.
-
Presenilins mediate a dual intramembranous gamma-secretase cleavage of Notch-1.EMBO J. 2002 Oct 15;21(20):5408-16. doi: 10.1093/emboj/cdf541. EMBO J. 2002. PMID: 12374741 Free PMC article.
-
NOTCH1 Signaling in Head and Neck Squamous Cell Carcinoma.Cells. 2020 Dec 12;9(12):2677. doi: 10.3390/cells9122677. Cells. 2020. PMID: 33322834 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

