Plant expansins are a complex multigene family with an ancient evolutionary origin
- PMID: 11891242
- PMCID: PMC152199
- DOI: 10.1104/pp.010658
Plant expansins are a complex multigene family with an ancient evolutionary origin
Abstract
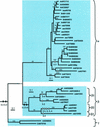
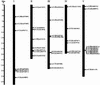
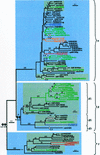
Expansins are a group of extracellular proteins that directly modify the mechanical properties of plant cell walls, leading to turgor-driven cell extension. Within the completely sequenced Arabidopsis genome, we identified 38 expansin sequences that fall into three discrete subfamilies. Based on phylogenetic analysis and shared intron patterns, we propose a new, systematic nomenclature of Arabidopsis expansins. Further phylogenetic analysis, including expansin sequences found here in monocots, pine (Pinus radiata, Pinus taeda), fern (Regnellidium diphyllum, Marsilea quadrifolia), and moss (Physcomitrella patens) indicate that the three plant expansin subfamilies arose and began diversifying very early in, if not before, colonization of land by plants. Closely related "expansin-like" sequences were also identified in the social amoeba, Dictyostelium discoidium, suggesting that these wall-modifying proteins have a very deep evolutionary origin.
Figures


Similar articles
-
Alpha-expansins in the semiaquatic ferns Marsilea quadrifolia and Regnellidium diphyllum: evolutionary aspects and physiological role in rachis elongation.Planta. 2000 Dec;212(1):85-92. doi: 10.1007/s004250000367. Planta. 2000. PMID: 11219587
-
Origin, Evolution, and Diversification of the Expansin Family in Plants.Int J Mol Sci. 2024 Nov 3;25(21):11814. doi: 10.3390/ijms252111814. Int J Mol Sci. 2024. PMID: 39519364 Free PMC article.
-
Portrait of the expansin superfamily in Physcomitrella patens: comparisons with angiosperm expansins.Ann Bot. 2007 Jun;99(6):1131-41. doi: 10.1093/aob/mcm044. Epub 2007 Apr 7. Ann Bot. 2007. PMID: 17416912 Free PMC article.
-
Expansins and cell growth.Curr Opin Plant Biol. 2003 Dec;6(6):603-10. doi: 10.1016/j.pbi.2003.09.003. Curr Opin Plant Biol. 2003. PMID: 14611960 Review.
-
Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs.J Plant Res. 2006 Jan;119(1):11-21. doi: 10.1007/s10265-005-0253-z. Epub 2006 Jan 13. J Plant Res. 2006. PMID: 16411016 Review.
Cited by
-
Plant expansins: diversity and interactions with plant cell walls.Curr Opin Plant Biol. 2015 Jun;25:162-72. doi: 10.1016/j.pbi.2015.05.014. Epub 2015 Jun 6. Curr Opin Plant Biol. 2015. PMID: 26057089 Free PMC article. Review.
-
Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening.J Exp Bot. 2011 Jan;62(2):815-23. doi: 10.1093/jxb/erq319. Epub 2010 Oct 19. J Exp Bot. 2011. PMID: 20959626 Free PMC article.
-
Purification and characterization of four beta-expansins (Zea m 1 isoforms) from maize pollen.Plant Physiol. 2003 Aug;132(4):2073-85. doi: 10.1104/pp.103.020024. Plant Physiol. 2003. PMID: 12913162 Free PMC article.
-
Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site.Plant Physiol. 2003 Mar;131(3):1178-90. doi: 10.1104/pp.016519. Plant Physiol. 2003. PMID: 12644669 Free PMC article.
-
Loosenin-Like Proteins from Phanerochaete carnosa Impact Both Cellulose and Chitin Fiber Networks.Appl Environ Microbiol. 2023 Jan 31;89(1):e0186322. doi: 10.1128/aem.01863-22. Epub 2023 Jan 16. Appl Environ Microbiol. 2023. PMID: 36645281 Free PMC article.
References
-
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–813. - PubMed
-
- Ceccardi TL, Barthe GA, Derrick KS. A novel protein associated with citrus blight has sequence similarity to expansin. Plant Mol Biol. 1998;38:775–783. - PubMed
-
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol. 1997;13:171–201. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

