Transcription activator interactions with multiple SWI/SNF subunits
- PMID: 11865042
- PMCID: PMC135607
- DOI: 10.1128/MCB.22.6.1615-1625.2002
Transcription activator interactions with multiple SWI/SNF subunits
Abstract
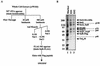
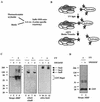
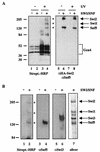
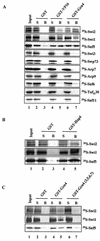
We have previously shown that the yeast SWI/SNF complex stimulates in vitro transcription from chromatin templates in an ATP-dependent manner. SWI/SNF function in this regard requires the presence of an activator with which it can interact directly, linking activator recruitment of SWI/SNF to transcriptional stimulation. In this study, we determine the SWI/SNF subunits that mediate its interaction with activators. Using a photo-cross-linking label transfer strategy, we show that the Snf5, Swi1, and Swi2/Snf2 subunits are contacted by the yeast acidic activators, Gcn4 and Hap4, in the context of the intact native SWI/SNF complex. In addition, we show that the same three subunits can interact individually with acidic activation domains, indicating that each subunit contributes to binding activators. Furthermore, mutations that reduce the activation potential of these activators also diminish its interaction with each of these SWI/SNF subunits. Thus, three distinct subunits of the SWI/SNF complex contribute to its interactions with activation domains.
Figures


Similar articles
-
Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA.Mol Cell Biol. 2003 Dec;23(23):8829-45. doi: 10.1128/MCB.23.23.8829-9945.2003. Mol Cell Biol. 2003. PMID: 14612422 Free PMC article.
-
Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays.Mol Cell. 1999 Oct;4(4):649-55. doi: 10.1016/s1097-2765(00)80216-6. Mol Cell. 1999. PMID: 10549297
-
Evidence that Swi/Snf directly represses transcription in S. cerevisiae.Genes Dev. 2002 Sep 1;16(17):2231-6. doi: 10.1101/gad.1009902. Genes Dev. 2002. PMID: 12208846 Free PMC article.
-
The mammalian SWI/SNF complex and the control of cell growth.Semin Cell Dev Biol. 1999 Apr;10(2):189-95. doi: 10.1006/scdb.1999.0300. Semin Cell Dev Biol. 1999. PMID: 10441072 Review.
-
Promoter targeting and chromatin remodeling by the SWI/SNF complex.Curr Opin Genet Dev. 2000 Apr;10(2):187-92. doi: 10.1016/s0959-437x(00)00068-x. Curr Opin Genet Dev. 2000. PMID: 10753786 Review.
Cited by
-
Interaction of papillomavirus E2 protein with the Brm chromatin remodeling complex leads to enhanced transcriptional activation.J Virol. 2007 Mar;81(5):2213-20. doi: 10.1128/JVI.01746-06. Epub 2006 Dec 6. J Virol. 2007. PMID: 17151122 Free PMC article.
-
A novel genetic circuitry governing hypoxic metabolic flexibility, commensalism and virulence in the fungal pathogen Candida albicans.PLoS Pathog. 2019 Dec 6;15(12):e1007823. doi: 10.1371/journal.ppat.1007823. eCollection 2019 Dec. PLoS Pathog. 2019. PMID: 31809527 Free PMC article.
-
SWI/SNF and the histone chaperone Rtt106 drive expression of the Pleiotropic Drug Resistance network genes.Nat Commun. 2022 Apr 12;13(1):1968. doi: 10.1038/s41467-022-29591-z. Nat Commun. 2022. PMID: 35413952 Free PMC article.
-
Role of chromatin during herpesvirus infections.Biochim Biophys Acta. 2009 Jun;1790(6):456-66. doi: 10.1016/j.bbagen.2009.03.019. Epub 2009 Mar 31. Biochim Biophys Acta. 2009. PMID: 19344747 Free PMC article. Review.
-
A novel mechanism for target gene-specific SWI/SNF recruitment via the Snf2p N-terminus.Nucleic Acids Res. 2011 May;39(10):4088-98. doi: 10.1093/nar/gkr004. Epub 2011 Jan 28. Nucleic Acids Res. 2011. PMID: 21278159 Free PMC article.
References
-
- Alley, S. C., A. D. Jones, P. Soumillion, and S. J. Benkovic. 1999. The carboxyl terminus of the bacteriophage T4 DNA polymerase contacts its sliding clamp at the subunit interface. J. Biol. Chem. 274:24485-24489. - PubMed
-
- Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. - PubMed
-
- Berger, S. L., W. D. Cress, A. Cress, S. J. Triezenberg, and L. Guarente. 1990. Selective inhibition of activated but not basal transcription by the acidic activtation domain of VP16: evidence for transcriptional adaptors. Cell 61:1199-1208. - PubMed
-
- Bochar, D. A., L. Wang, H. Beniya, A. Kinev, Y. Xue, W. S. Lane, W. Wang, F. Kashanchi, and R. Shiekhattar. 2000. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102:257-265. - PubMed
-
- Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
