Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit
- PMID: 11826110
- PMCID: PMC6758529
- DOI: 10.1523/JNEUROSCI.22-03-00803.2002
Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit
Abstract
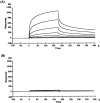
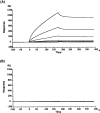
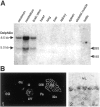
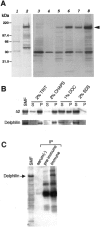
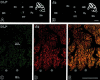
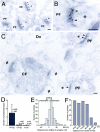
The glutamate receptor delta2 (GluRdelta2) subunit is selectively expressed in cerebellar Purkinje cells and plays an important role in cerebellar long-term depression, motor learning, motor coordination, and synapse development. We identified a novel GluRdelta2-interacting protein, named Delphilin, that contains a single PDZ domain and formin homology (FH) domains FH1 and FH2 plus coiled-coil structure. As far as we know, this is the first reported protein that contains both PDZ and FH domains. Yeast two-hybrid and surface plasmon resonance (SPR) analyses indicated that Delphilin interacts with the GluRdelta2 C terminus via its PDZ domain. This was also supported by coimmunoprecipitation experiments using a heterologous expression system in mammalian cells. Yeast cell and SPR analyses also demonstrated the possibility that the FH1 proline-rich region of Delphilin interacts with profilin, an actin-binding protein, and with the Src homology 3 domain of neuronal Src protein tyrosine kinase. In situ hybridization demonstrated the highest expression of Delphilin mRNA in Purkinje cells. Delphilin polypeptide was highly enriched in the synaptosomal membrane fraction of the cerebellum and coimmunoprecipitated with the GluRdelta2 subunit. The post-embedding immunogold technique demonstrated that Delphilin is selectively localized at the postsynaptic junction site of the parallel fiber-Purkinje cell synapse and colocalized with GluRdelta2. Thus, Delphilin is a postsynaptic scaffolding protein at the parallel fiber-Purkinje cell synapse, where it may serve to link GluRdelta2 with actin cytoskeleton and various signaling molecules.
Figures



Similar articles
-
Identification and characterization of a novel Delphilin variant with an alternative N-terminus.Brain Res Mol Brain Res. 2005 Nov 18;141(1):83-94. doi: 10.1016/j.molbrainres.2005.08.006. Epub 2005 Sep 15. Brain Res Mol Brain Res. 2005. PMID: 16168524
-
Binding of glutamate receptor delta2 to its scaffold protein, Delphilin, is regulated by PKA.Biochem Biophys Res Commun. 2006 Nov 24;350(3):748-52. doi: 10.1016/j.bbrc.2006.09.109. Epub 2006 Sep 28. Biochem Biophys Res Commun. 2006. PMID: 17027646
-
The synaptic scaffolding protein Delphilin interacts with monocarboxylate transporter 2.Neuroreport. 2007 Mar 26;18(5):489-93. doi: 10.1097/WNR.0b013e3280586821. Neuroreport. 2007. PMID: 17496809
-
Glutamate-receptor-like molecule GluRδ2 involved in synapse formation at parallel fiber-Purkinje neuron synapses.Cerebellum. 2012 Mar;11(1):71-7. doi: 10.1007/s12311-010-0170-0. Cerebellum. 2012. PMID: 20387025 Review.
-
An orphan ionotropic glutamate receptor: the delta2 subunit.Neuroscience. 2009 Jan 12;158(1):67-77. doi: 10.1016/j.neuroscience.2008.02.050. Epub 2008 Mar 6. Neuroscience. 2009. PMID: 18424007 Review.
Cited by
-
Actin cytoskeleton in dendritic spine development and plasticity.Curr Opin Neurobiol. 2016 Aug;39:86-92. doi: 10.1016/j.conb.2016.04.010. Epub 2016 Apr 30. Curr Opin Neurobiol. 2016. PMID: 27138585 Free PMC article. Review.
-
Neuronal profilin isoforms are addressed by different signalling pathways.PLoS One. 2012;7(3):e34167. doi: 10.1371/journal.pone.0034167. Epub 2012 Mar 28. PLoS One. 2012. PMID: 22470532 Free PMC article.
-
Glutamate receptor δ2 associates with metabotropic glutamate receptor 1 (mGluR1), protein kinase Cγ, and canonical transient receptor potential 3 and regulates mGluR1-mediated synaptic transmission in cerebellar Purkinje neurons.J Neurosci. 2012 Oct 31;32(44):15296-308. doi: 10.1523/JNEUROSCI.0705-12.2012. J Neurosci. 2012. PMID: 23115168 Free PMC article.
-
Novel espin actin-bundling proteins are localized to Purkinje cell dendritic spines and bind the Src homology 3 adapter protein insulin receptor substrate p53.J Neurosci. 2003 Feb 15;23(4):1310-9. doi: 10.1523/JNEUROSCI.23-04-01310.2003. J Neurosci. 2003. PMID: 12598619 Free PMC article.
-
Role of actin cytoskeleton in the organization and function of ionotropic glutamate receptors.Curr Res Struct Biol. 2021 Oct 14;3:277-289. doi: 10.1016/j.crstbi.2021.10.001. eCollection 2021. Curr Res Struct Biol. 2021. PMID: 34766008 Free PMC article. Review.
References
-
- Adamson JG, Zhou NE, Hodges RS. Structure, function and application of the coiled-coil protein folding motif. Curr Opin Biotechnol. 1993;4:428–437. - PubMed
-
- Aoki C, Miko I, Oviedo H, Mikelandze-Dvali T, Alexandre L, Sweeney N, Bredt DS. Electron microscopic immunocytochemical detection of PSD-95, PSD-93, SAP-102, and SAP-97 at postsynaptic, presynaptic, and nonsynaptic sites of adult and neonatal rat visual cortex. Synapse. 2001;40:239–257. - PubMed
-
- Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M. Selective expression of the glutamate receptor channel δ2 subunit in cerebellar Purkinje cells. Biochem Biophys Res Commun. 1993;197:1267–1276. - PubMed
-
- Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. - PubMed
-
- Blackstone CD, Moss SJ, Martin LJ, Levey AI, Price DL, Hugani RL. Biochemical characterization and localization of a non-N-methyl-d-aspartate glutamate receptor in rat brain. J Neurochem. 1992;58:1118–1126. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
