NF1/X represses PDGF A-chain transcription by interacting with Sp1 and antagonizing Sp1 occupancy of the promoter
- PMID: 11823426
- PMCID: PMC125828
- DOI: 10.1093/emboj/21.3.334
NF1/X represses PDGF A-chain transcription by interacting with Sp1 and antagonizing Sp1 occupancy of the promoter
Abstract
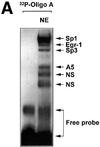
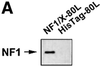
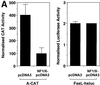
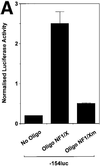
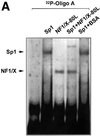
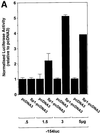
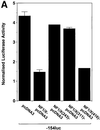
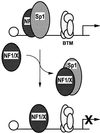
The regulatory mechanisms mediating basal and inducible platelet-derived growth factor (PDGF)-A expression have been the focus of intense recent investigation, but repression of PDGF-A expression is largely unexplored. Here we isolated a nuclear factor that interacts with the proximal region of the PDGF-A promoter using bulk binding assays and chromatography techniques. Peptide mass fingerprint and supershift analysis revealed this DNA-binding protein to be NF1/X. NF1/X repressed PDGF-A promoter-dependent transcription and endogenous mRNA expression, which was reversible by oligonucleotide decoys bearing an NF1/X-binding site. Mutation in the DNA-binding domain of NF1/X abolished its repression of PDGF-A promoter. NF1/X antagonized the activity of a known activator of the PDGF-A chain, Sp1, by inhibiting its occupancy of the proximal PDGF-A promoter. NF1/X physically and specifically interacts with Sp1 via its subtype-specific domain and blocks Sp1 induction of the promoter. NF1/X residues 311-416 mediated NF1/X suppression of basal PDGF-A transcription, whereas residues 243-416 were required for NF1/X repression of Sp1-inducible promoter activity. These findings demonstrate that repression of PDGF-A gene transcription is governed by interplay between NF1/X and Sp1.
Figures

















Similar articles
-
Ets-1 stimulates platelet-derived growth factor A-chain gene transcription and vascular smooth muscle cell growth via cooperative interactions with Sp1.Circ Res. 2004 Sep 3;95(5):479-87. doi: 10.1161/01.RES.0000141135.36279.67. Epub 2004 Aug 5. Circ Res. 2004. PMID: 15297375
-
Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts.Exp Cell Res. 2005 Jan 1;302(1):129-42. doi: 10.1016/j.yexcr.2004.08.028. Exp Cell Res. 2005. PMID: 15541732
-
A nuclear factor other than Sp1 binds the GC-rich promoter of the gene encoding rat poly(ADP-ribose) polymerase in vitro.Biochem Cell Biol. 1997;75(4):427-34. Biochem Cell Biol. 1997. PMID: 9493965
-
Transcription of the platelet-derived growth factor A-chain gene.Cytokine Growth Factor Rev. 2003 Oct;14(5):427-46. doi: 10.1016/s1359-6101(03)00051-0. Cytokine Growth Factor Rev. 2003. PMID: 12948525 Review.
-
S1-nuclease-sensitive DNA structures contribute to transcriptional regulation of the human PDGF A-chain.Prog Nucleic Acid Res Mol Biol. 1996;55:227-44. doi: 10.1016/s0079-6603(08)60195-6. Prog Nucleic Acid Res Mol Biol. 1996. PMID: 8787612 Review. No abstract available.
Cited by
-
High levels of glucose induce "metabolic memory" in cardiomyocyte via epigenetic histone H3 lysine 9 methylation.Mol Biol Rep. 2012 Sep;39(9):8891-8. doi: 10.1007/s11033-012-1756-z. Epub 2012 Jun 17. Mol Biol Rep. 2012. PMID: 22707199
-
Nuclear factor I-C links platelet-derived growth factor and transforming growth factor beta1 signaling to skin wound healing progression.Mol Cell Biol. 2009 Nov;29(22):6006-17. doi: 10.1128/MCB.01921-08. Epub 2009 Sep 14. Mol Cell Biol. 2009. PMID: 19752192 Free PMC article.
-
Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans.Neuropsychopharmacology. 2013 Jan;38(1):111-23. doi: 10.1038/npp.2012.149. Epub 2012 Sep 12. Neuropsychopharmacology. 2013. PMID: 22968814 Free PMC article. Review.
-
Control of renin [corrected] gene expression.Pflugers Arch. 2013 Jan;465(1):13-21. doi: 10.1007/s00424-012-1110-2. Epub 2012 May 11. Pflugers Arch. 2013. PMID: 22576577 Free PMC article. Review.
-
CCR5 Promoter Polymorphisms Associated With Pulmonary Tuberculosis in a Chinese Han Population.Front Immunol. 2021 Feb 19;11:544548. doi: 10.3389/fimmu.2020.544548. eCollection 2020. Front Immunol. 2021. PMID: 33679683 Free PMC article.
References
-
- Adams A.D., Choate,D.M. and Thompson,M. (1995) NF1-L is the DNA-binding component of the protein complex at the peripherin negative regulatory element. J. Biol. Chem., 270, 6975–6983. - PubMed
-
- Bergsten E., Uutela,M., Li,X., Pietras,K., Ostman,A., Heldin,C.-H., Alitalo,K. and Eriksson,U. (2001) PDGF-D is a specific, protease-activated ligand for the PDGF β-receptor. Nature Cell Biol., 3, 512–516. - PubMed
-
- Blomquist P., Li,Q. and Wrange,O. (1996) The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J. Biol. Chem., 271, 153–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

